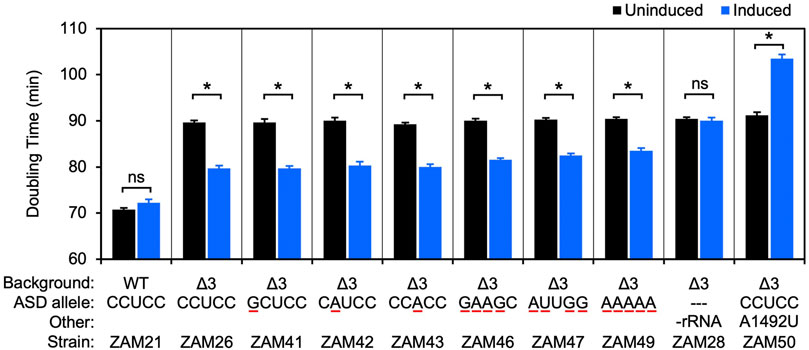
Accessibility of the Shine-Dalgarno Sequence Dictates N-Terminal Codon Bias in E. coli - ScienceDirect

Influences on gene expression in vivo by a Shine–Dalgarno sequence - Jin - 2006 - Molecular Microbiology - Wiley Online Library
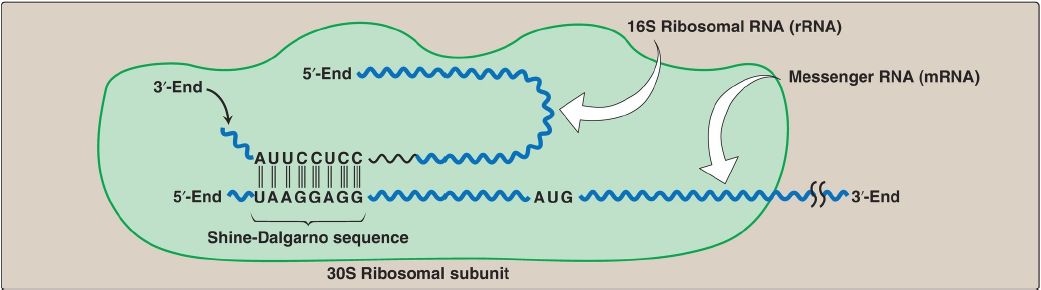
![PDF] Shine-Dalgarno Anti-Shine-Dalgarno Sequence Interactions and Their Functional Role in Translational Efficiency of Bacteria and Archaea | Semantic Scholar PDF] Shine-Dalgarno Anti-Shine-Dalgarno Sequence Interactions and Their Functional Role in Translational Efficiency of Bacteria and Archaea | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/ca6e3ce301334ffc5d3a7df6b8a8e5d2bc19ed16/23-Figure1.2-1.png)
PDF] Shine-Dalgarno Anti-Shine-Dalgarno Sequence Interactions and Their Functional Role in Translational Efficiency of Bacteria and Archaea | Semantic Scholar
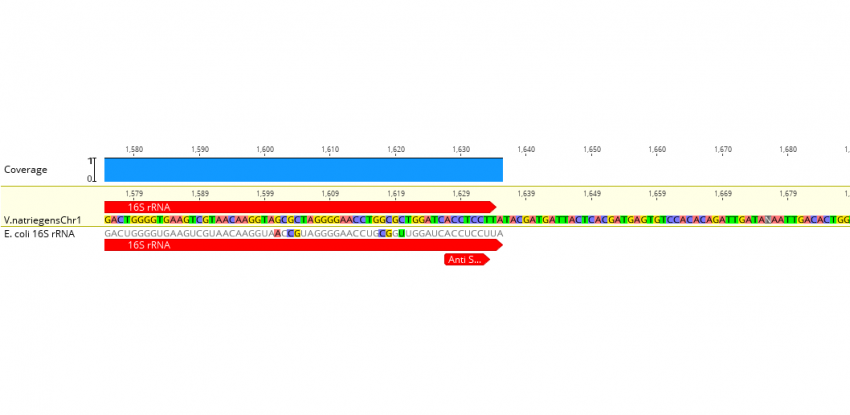
Leveraging genome-wide datasets to quantify the functional role of the anti- Shine–Dalgarno sequence in regulating translation efficiency - Publications - Amaral Lab

Extending the Spacing between the Shine–Dalgarno Sequence and P-Site Codon Reduces the Rate of mRNA Translocation - ScienceDirect
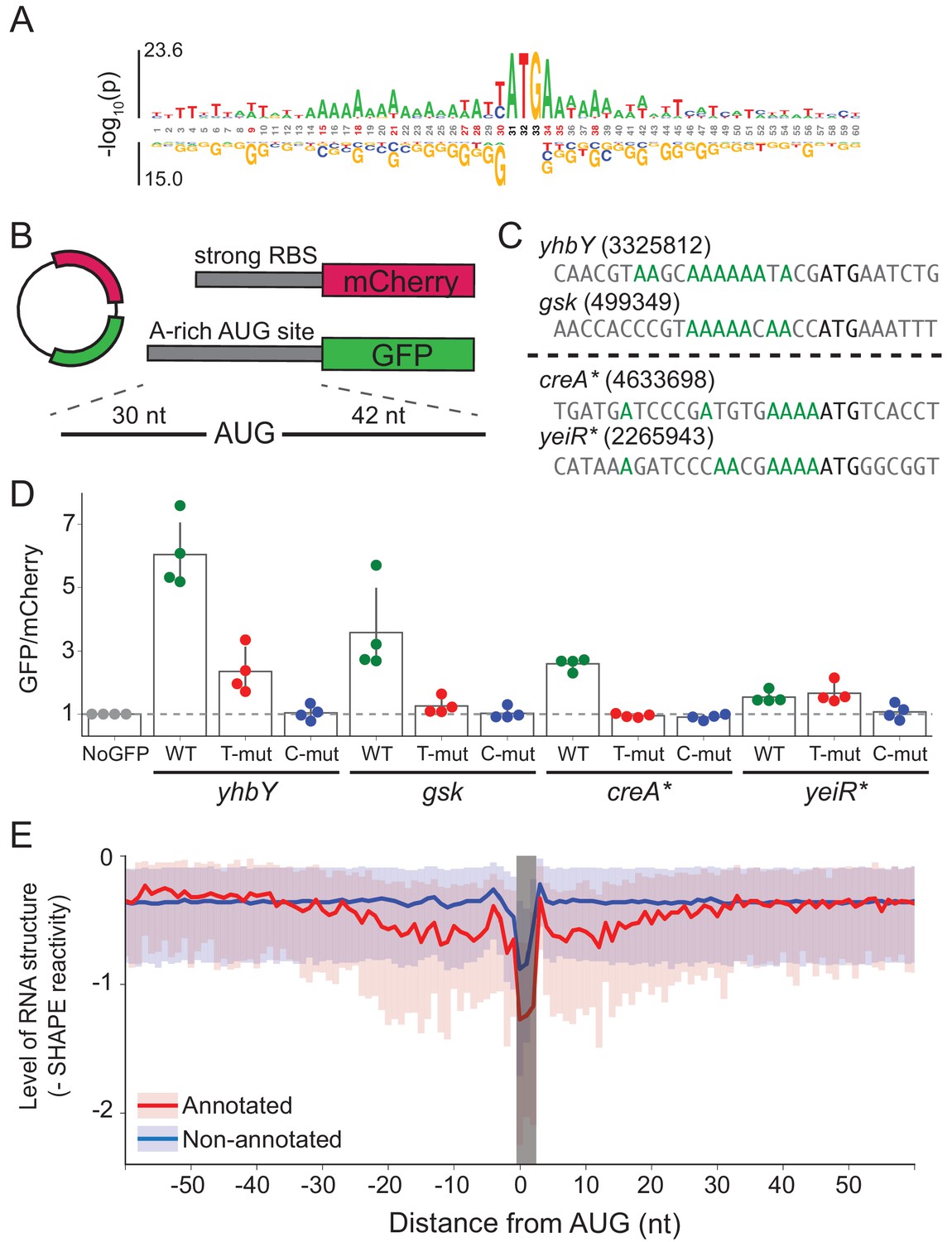
Translational initiation in E. coli occurs at the correct sites genome-wide in the absence of mRNA-rRNA base-pairing | eLife

Silent mutations in the Shine-Dalgarno-like sequence and the predicted... | Download Scientific Diagram

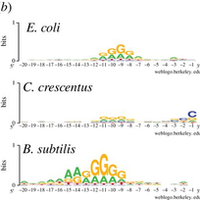
Elucidating the 16S rRNA 3′ boundaries and defining optimal SD/aSD pairing in Escherichia coli and Bacillus subtilis using RNA-Seq data | Scientific Reports

What is the Difference Between Shine Dalgarno and Kozak Sequence | Compare the Difference Between Similar Terms
Re-annotation of 12,495 prokaryotic 16S rRNA 3' ends and analysis of Shine- Dalgarno and anti-Shine-Dalgarno sequences | PLOS ONE

An extended Shine–Dalgarno sequence in mRNA functionally bypasses a vital defect in initiator tRNA | PNAS

Diversity of translation initiation mechanisms across bacterial species is driven by environmental conditions and growth demands | bioRxiv

A Snapshot of the 30S Ribosomal Subunit Capturing mRNA via the Shine- Dalgarno Interaction: Structure
![PDF] Shine-Dalgarno Anti-Shine-Dalgarno Sequence Interactions and Their Functional Role in Translational Efficiency of Bacteria and Archaea | Semantic Scholar PDF] Shine-Dalgarno Anti-Shine-Dalgarno Sequence Interactions and Their Functional Role in Translational Efficiency of Bacteria and Archaea | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/ca6e3ce301334ffc5d3a7df6b8a8e5d2bc19ed16/21-Figure1.1-1.png)
PDF] Shine-Dalgarno Anti-Shine-Dalgarno Sequence Interactions and Their Functional Role in Translational Efficiency of Bacteria and Archaea | Semantic Scholar