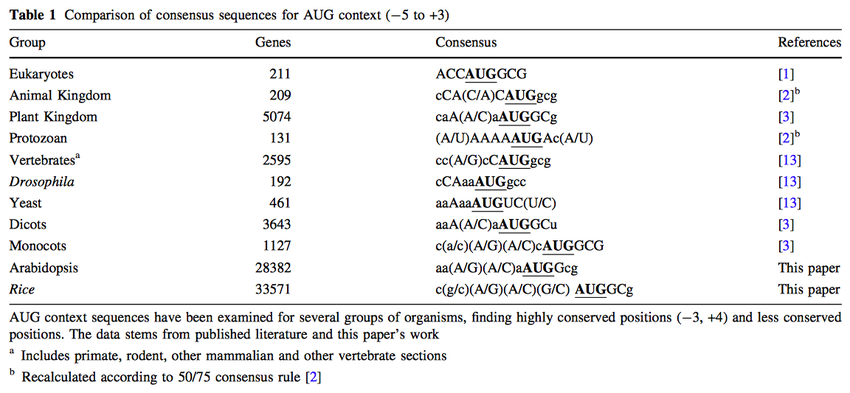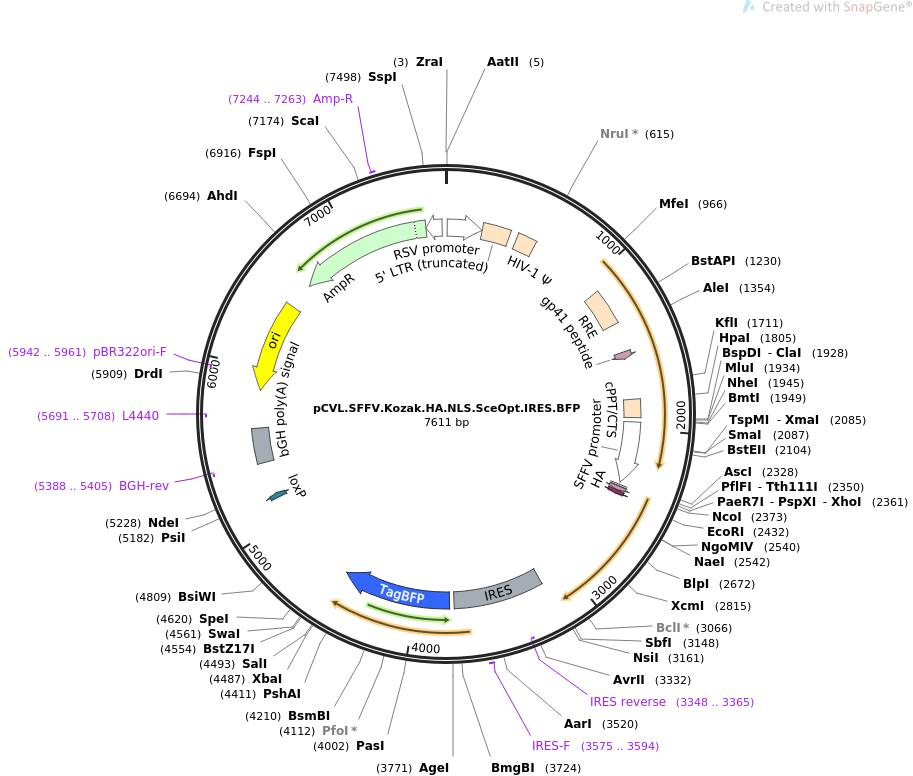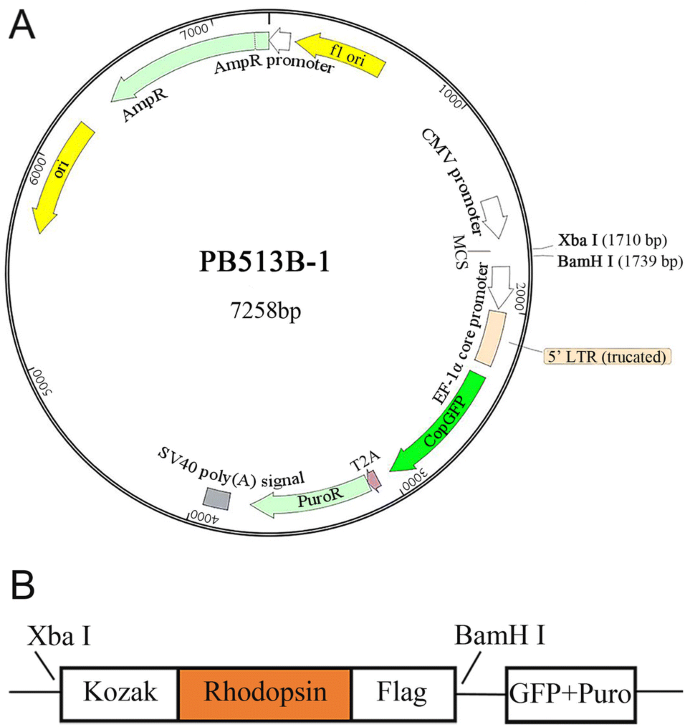
Heterologous expression and cell membrane localization of dinoflagellate opsins (rhodopsin proteins) in mammalian cells | SpringerLink
Alternative Transcription Initiation and the AUG Context Configuration Control Dual-Organellar Targeting and Functional Competen
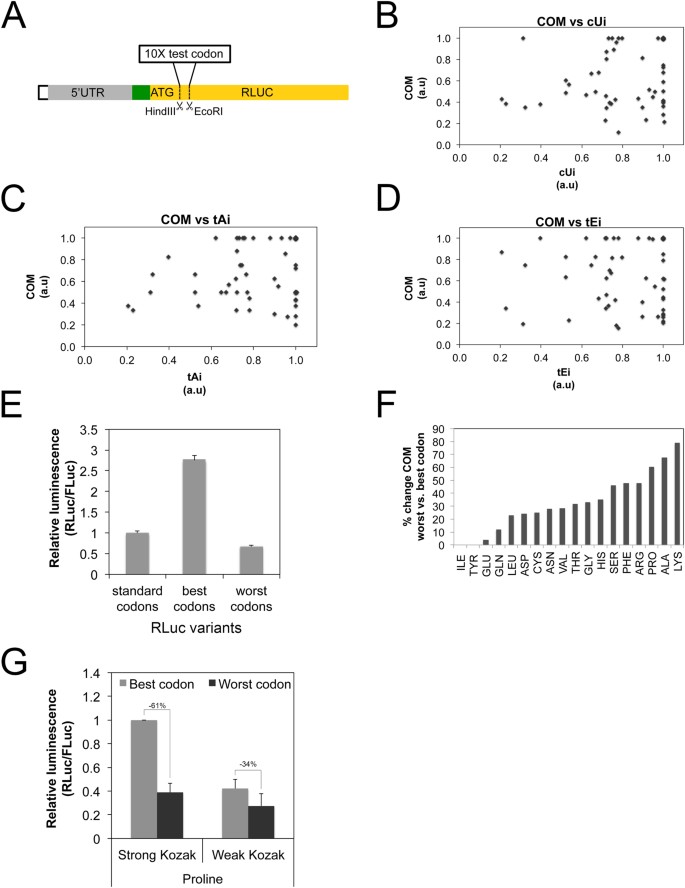
Changes in global translation elongation or initiation rates shape the proteome via the Kozak sequence | Scientific Reports

Kozak sequence improved the expression of 2A downstream gene by its... | Download Scientific Diagram
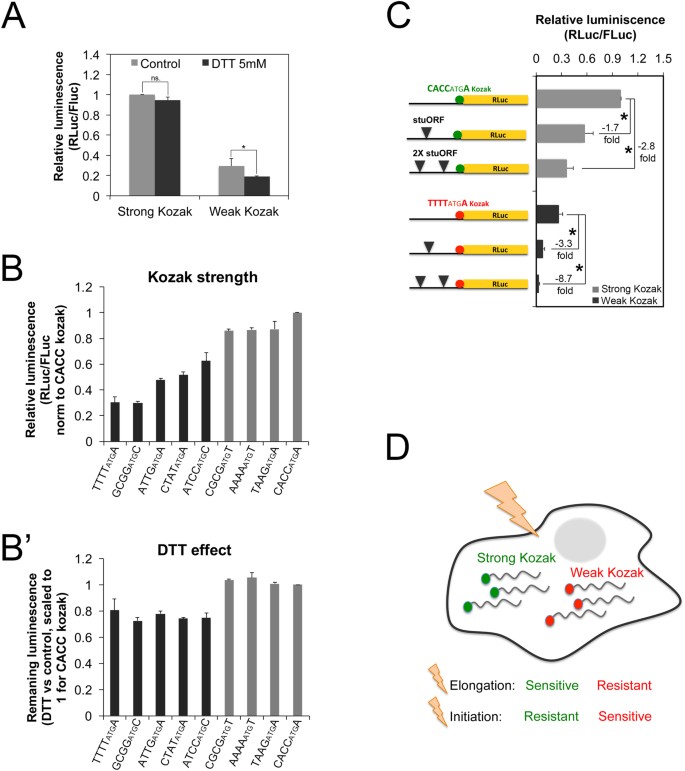
Changes in global translation elongation or initiation rates shape the proteome via the Kozak sequence | Scientific Reports

Fine-tuning the expression of pathway gene in yeast using a regulatory library formed by fusing a synthetic minimal promoter with different Kozak variants | Microbial Cell Factories | Full Text
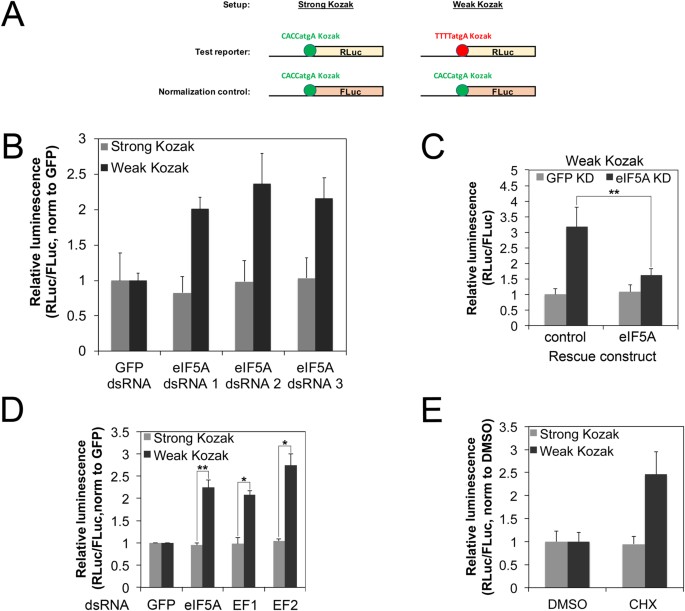
Changes in global translation elongation or initiation rates shape the proteome via the Kozak sequence | Scientific Reports
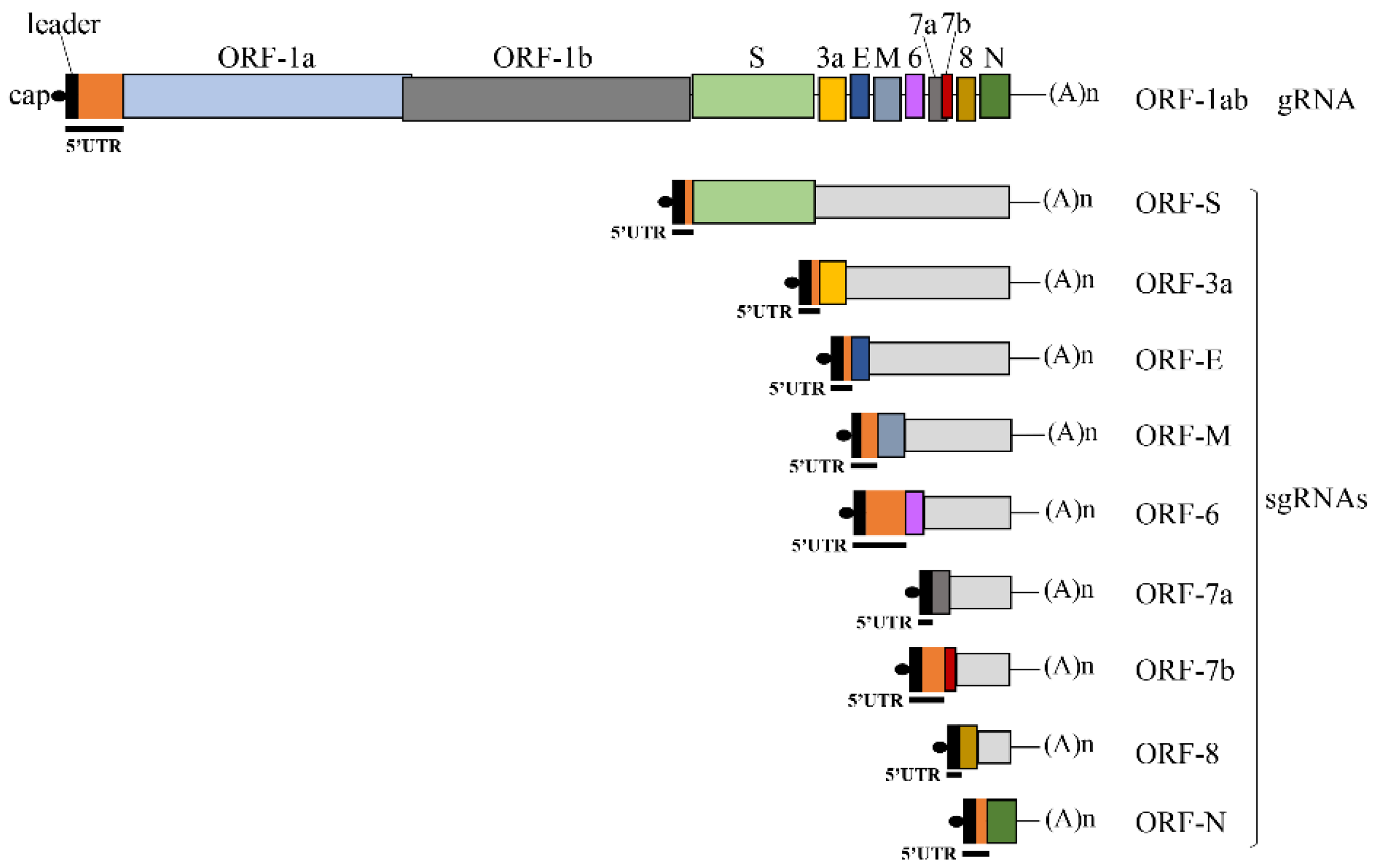
Viruses | Free Full-Text | Translation of SARS-CoV-2 gRNA Is Extremely Efficient and Competitive despite a High Degree of Secondary Structures and the Presence of an uORF
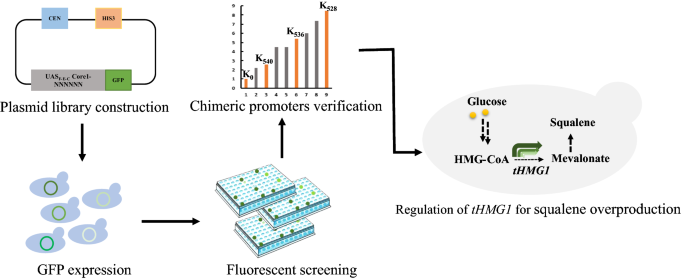
Fine-tuning the expression of pathway gene in yeast using a regulatory library formed by fusing a synthetic minimal promoter with different Kozak variants | Microbial Cell Factories | Full Text
The +4G Site in Kozak Consensus Is Not Related to the Efficiency of Translation Initiation | PLOS ONE

Structural insights into the Kozak sequence interactions within the mammalian late-stage initiation complexes | bioRxiv