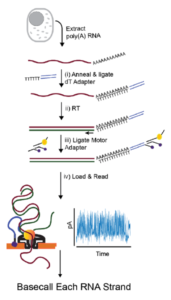
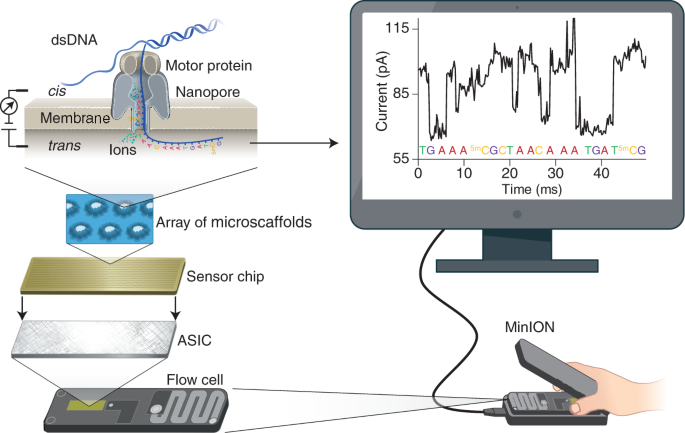
MasterOfPores: A Workflow for the Analysis of Oxford Nanopore Direct RNA Sequencing Datasets | Semantic Scholar

Improved nanopore direct RNA sequencing of cardiac myocyte samples by selective mt-RNA depletion - Journal of Molecular and Cellular Cardiology

Barcoding and demultiplexing Oxford Nanopore native RNA sequencing reads with deep residual learning | bioRxiv

MasterOfPores: A Workflow for the Analysis of Oxford Nanopore Direct RNA Sequencing Datasets | Semantic Scholar
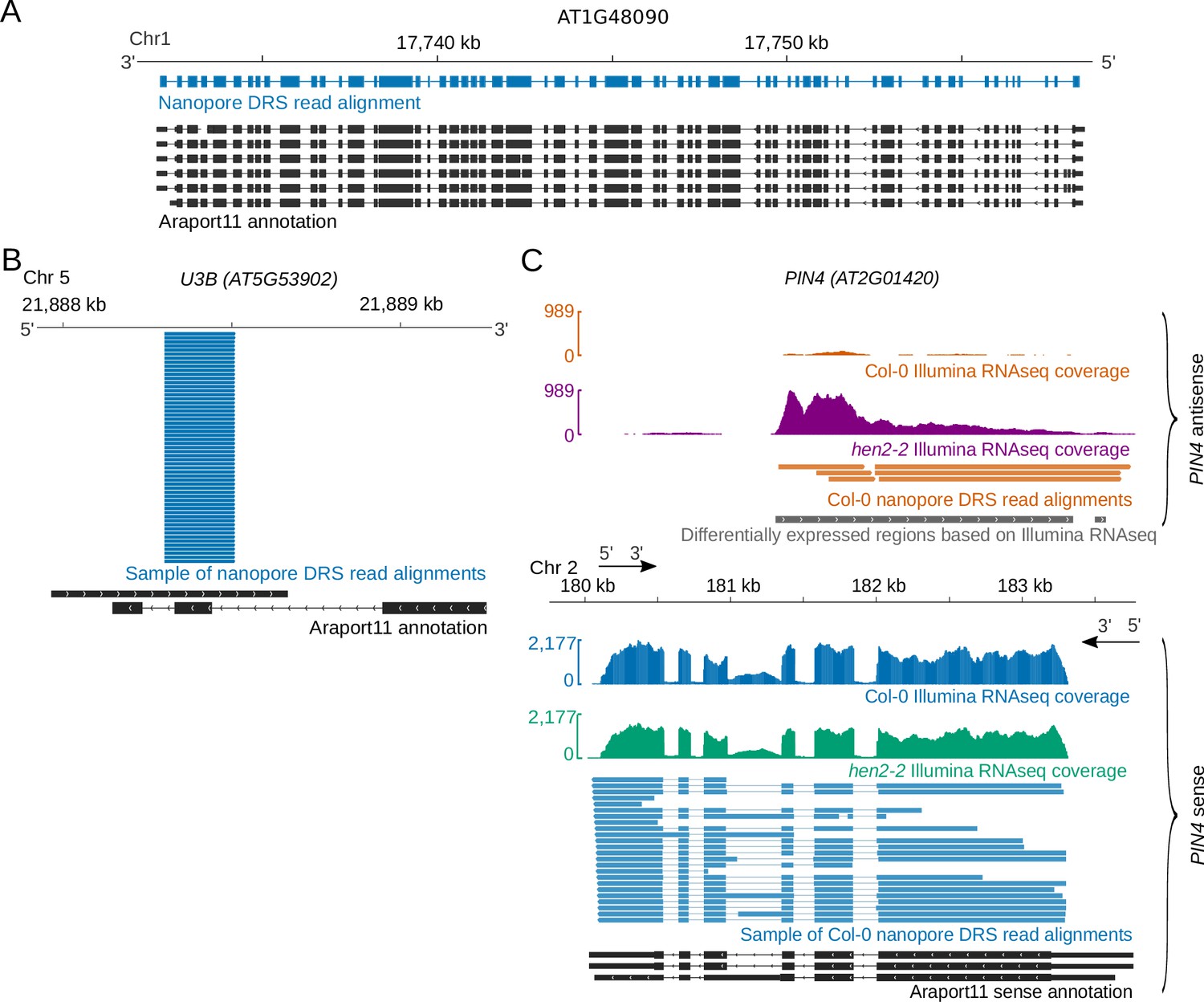
Nanopore direct RNA sequencing maps the complexity of Arabidopsis mRNA processing and m6A modification | eLife

Nanopore-based native RNA sequencing provides insights into prokaryotic transcription, operon structures, rRNA maturation and modifications | bioRxiv

Direct detection of RNA modifications and structure using single molecule nanopore sequencing | bioRxiv