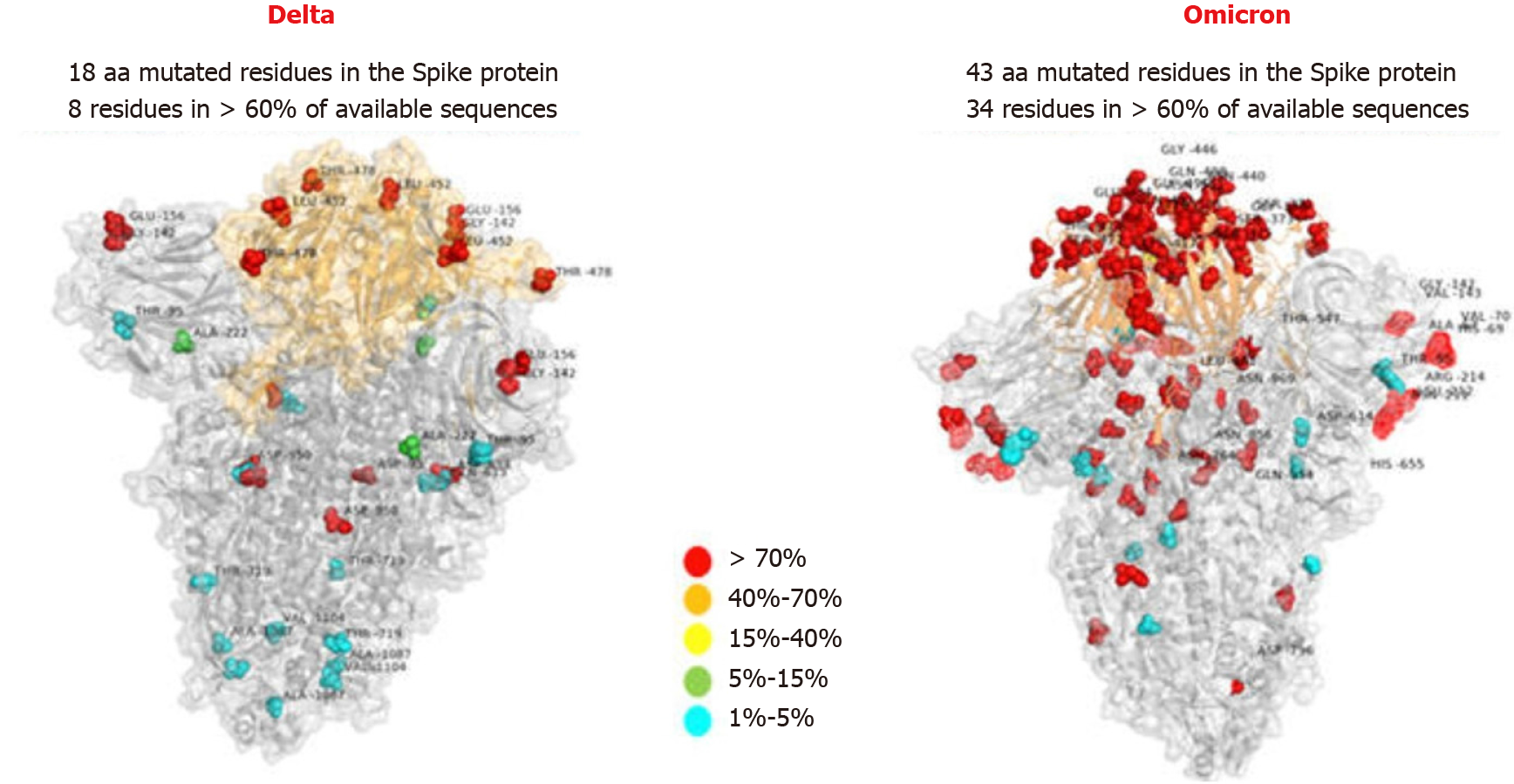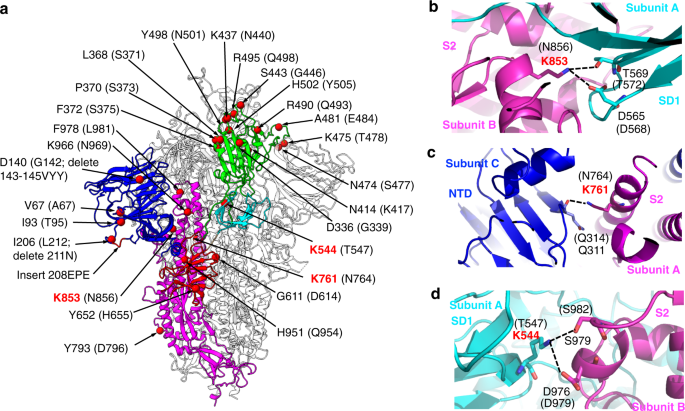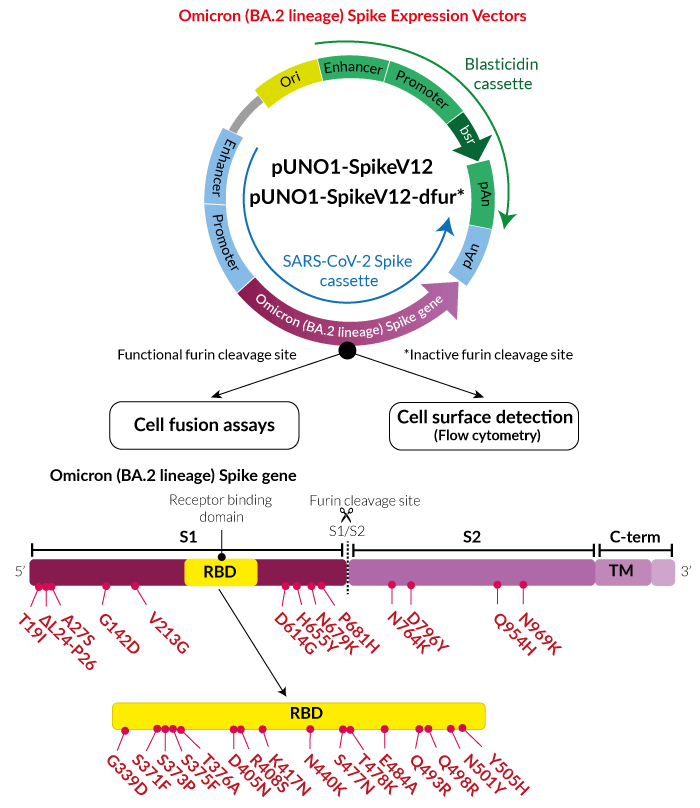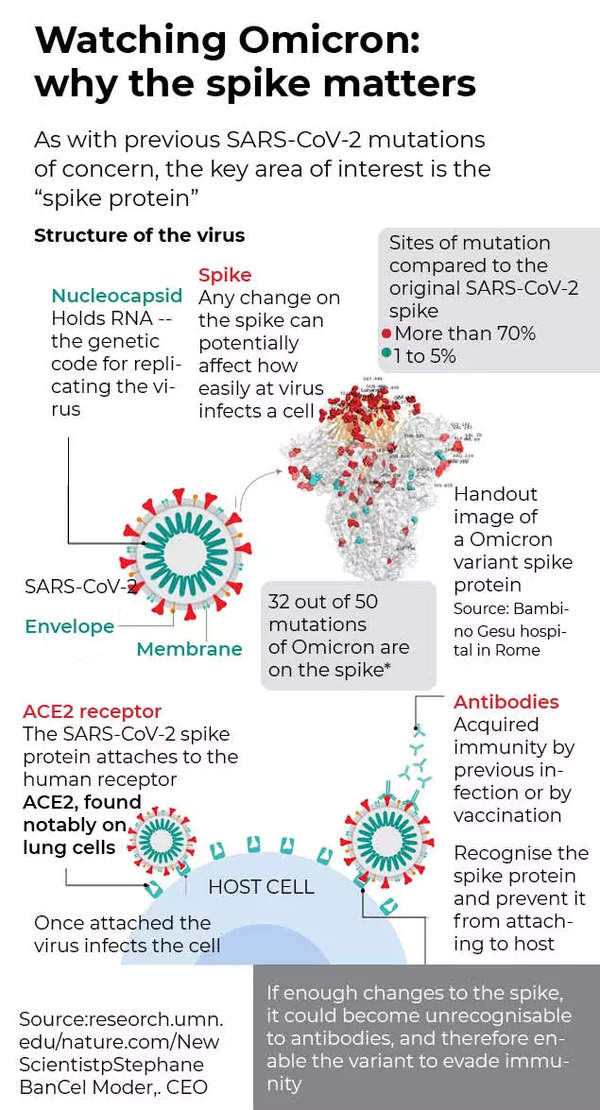
Frontiers | Omicron: A Heavily Mutated SARS-CoV-2 Variant Exhibits Stronger Binding to ACE2 and Potently Escapes Approved COVID-19 Therapeutic Antibodies

Omicron mutations enhance infectivity and reduce antibody neutralization of SARS-CoV-2 virus-like particles | PNAS
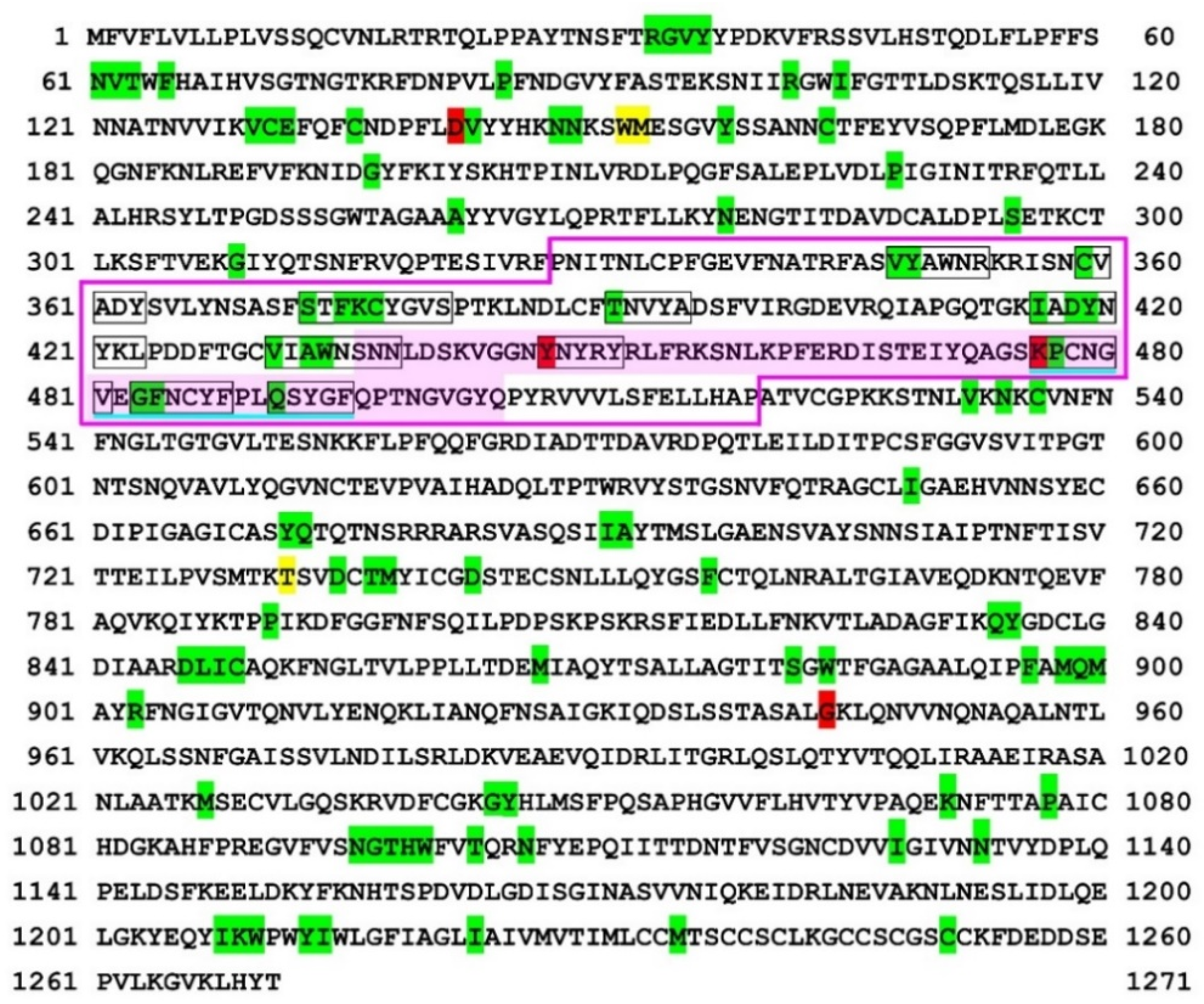
COVID | Free Full-Text | Nonself Mutations in the Spike Protein Suggest an Increase in the Antigenicity and a Decrease in the Virulence of the Omicron Variant of SARS-CoV-2

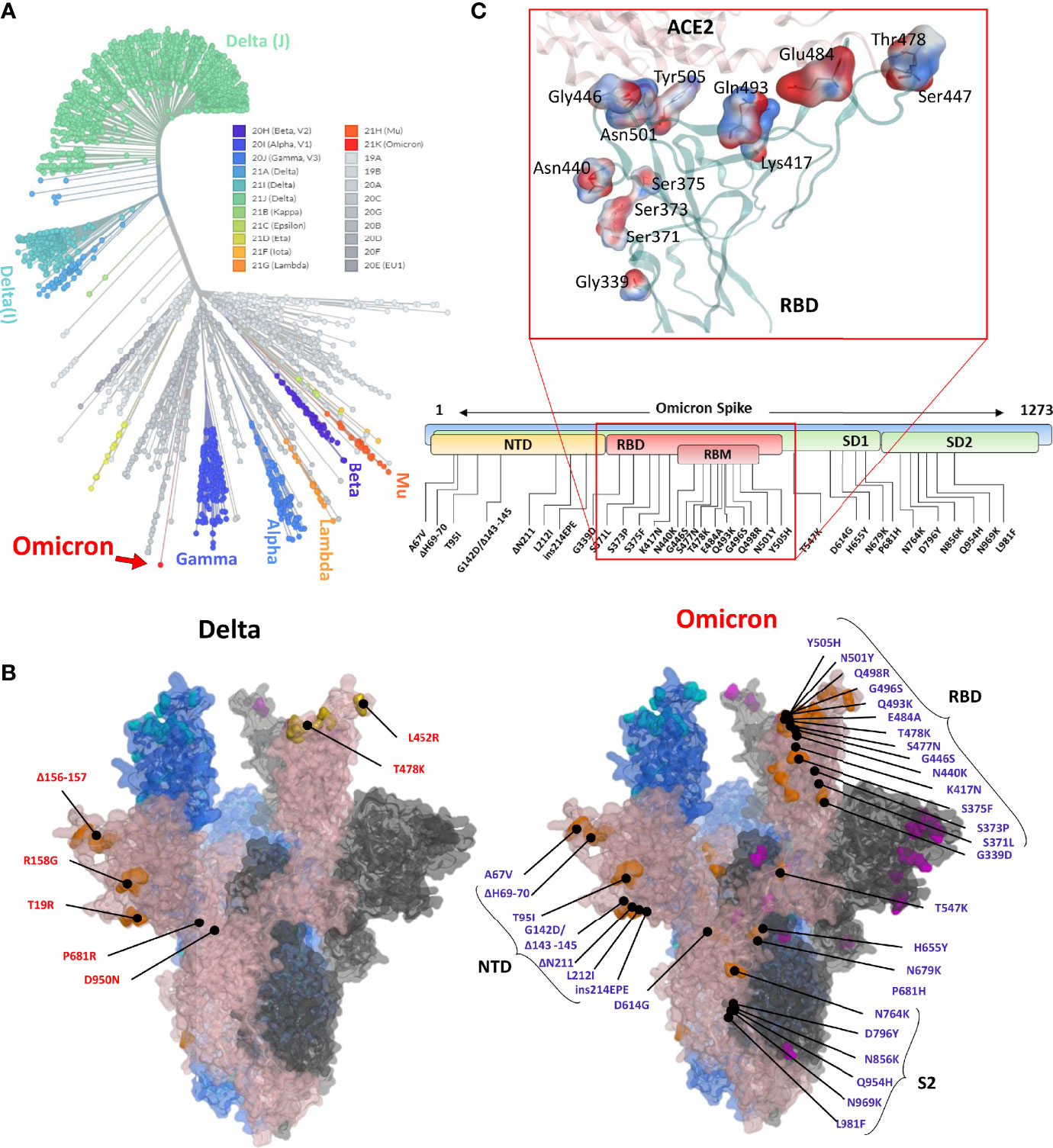
SARS-CoV-2 Omicron variant: Antibody evasion and cryo-EM structure of spike protein–ACE2 complex | Science

Interaction Analysis of the Spike Protein of Delta and Omicron Variants of SARS-CoV-2 with hACE2 and Eight Monoclonal Antibodies Using the Fragment Molecular Orbital Method | Journal of Chemical Information and Modeling
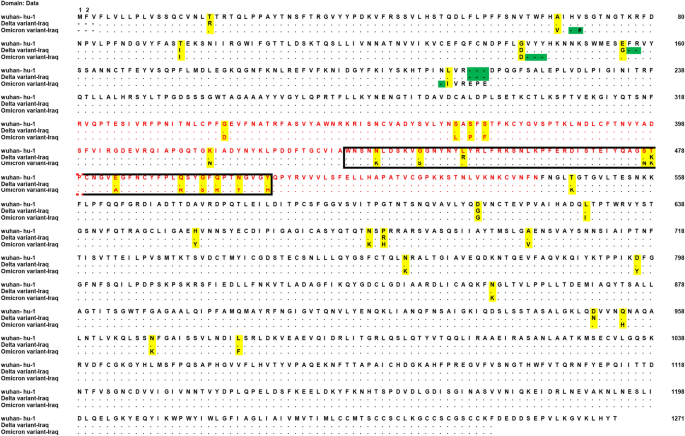
Molecular and computational analysis of spike protein of newly emerged omicron variant in comparison to the delta variant of SARS-CoV-2 in Iraq | SpringerLink
Frontiers | Predictions of the SARS-CoV-2 Omicron Variant (B.1.1.529) Spike Protein Receptor-Binding Domain Structure and Neutralizing Antibody Interactions
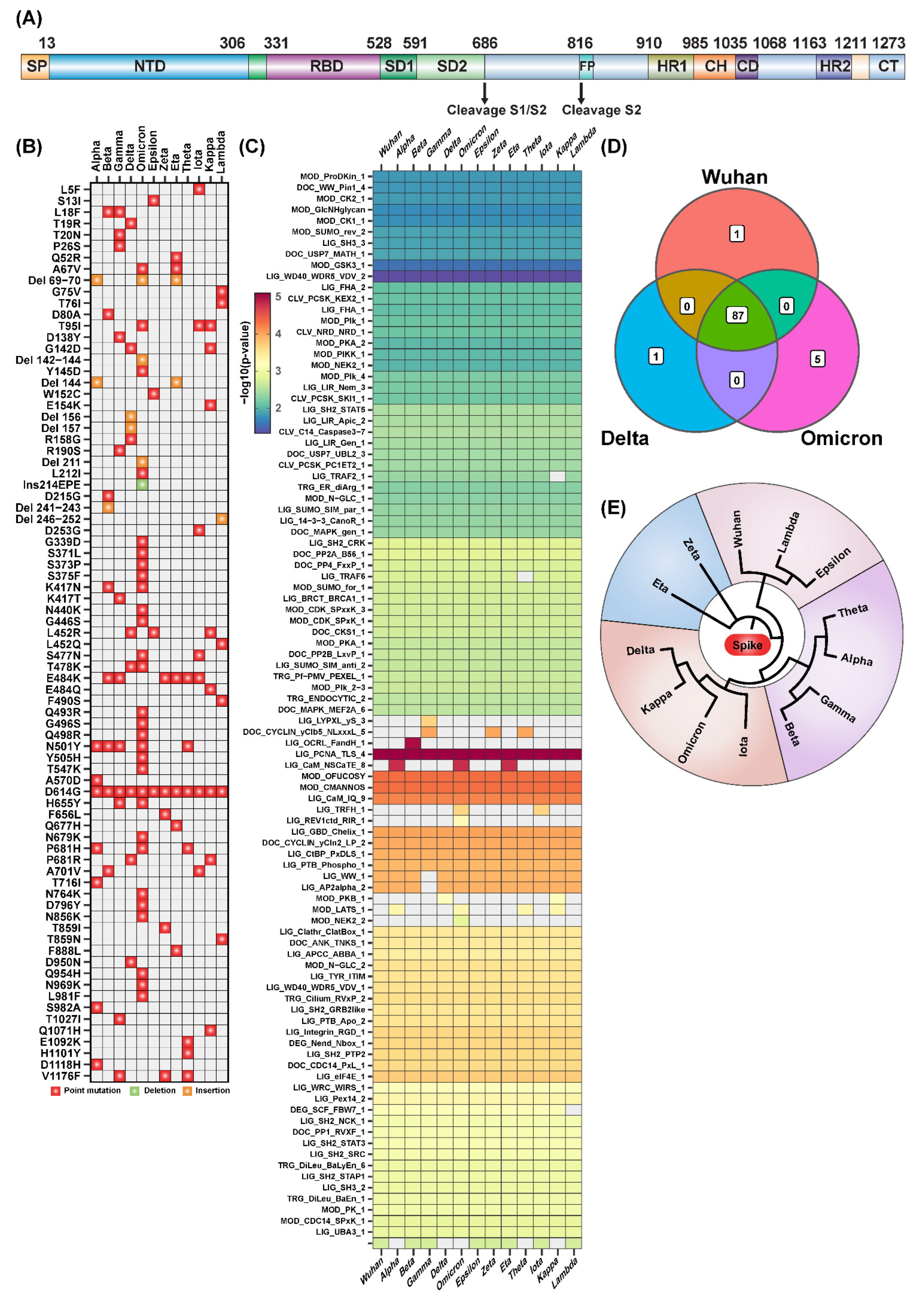
IJMS | Free Full-Text | Computational Analysis of Short Linear Motifs in the Spike Protein of SARS-CoV-2 Variants Provides Possible Clues into the Immune Hijack and Evasion Mechanisms of Omicron Variant
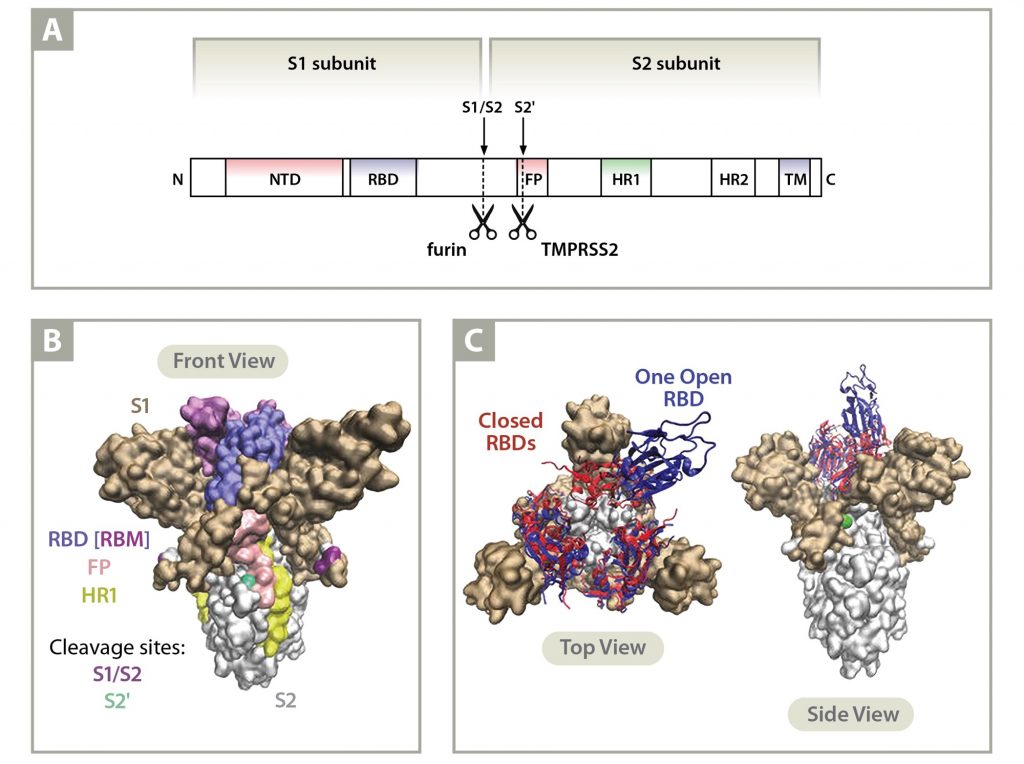
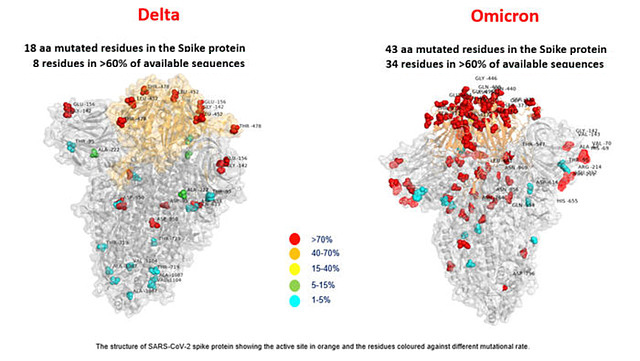
Implications of the Mutations in the Spike Protein of the Omicron Variant of Concern (VoC) of SARS-CoV-2 – Signature Science
Determinants of Spike infectivity, processing, and neutralization in SARS-CoV-2 Omicron subvariants BA.1 and BA.2
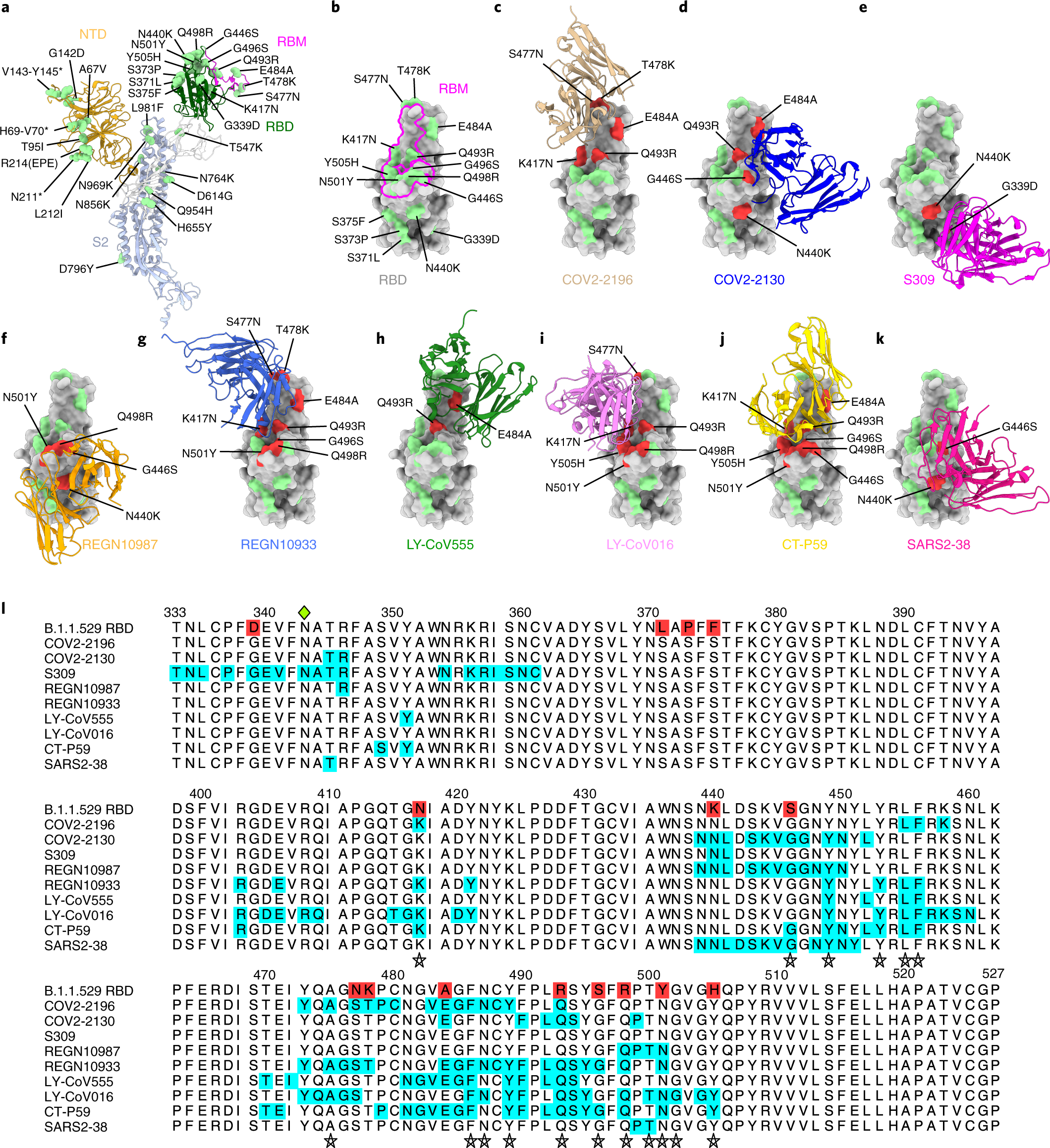
An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by therapeutic monoclonal antibodies | Nature Medicine
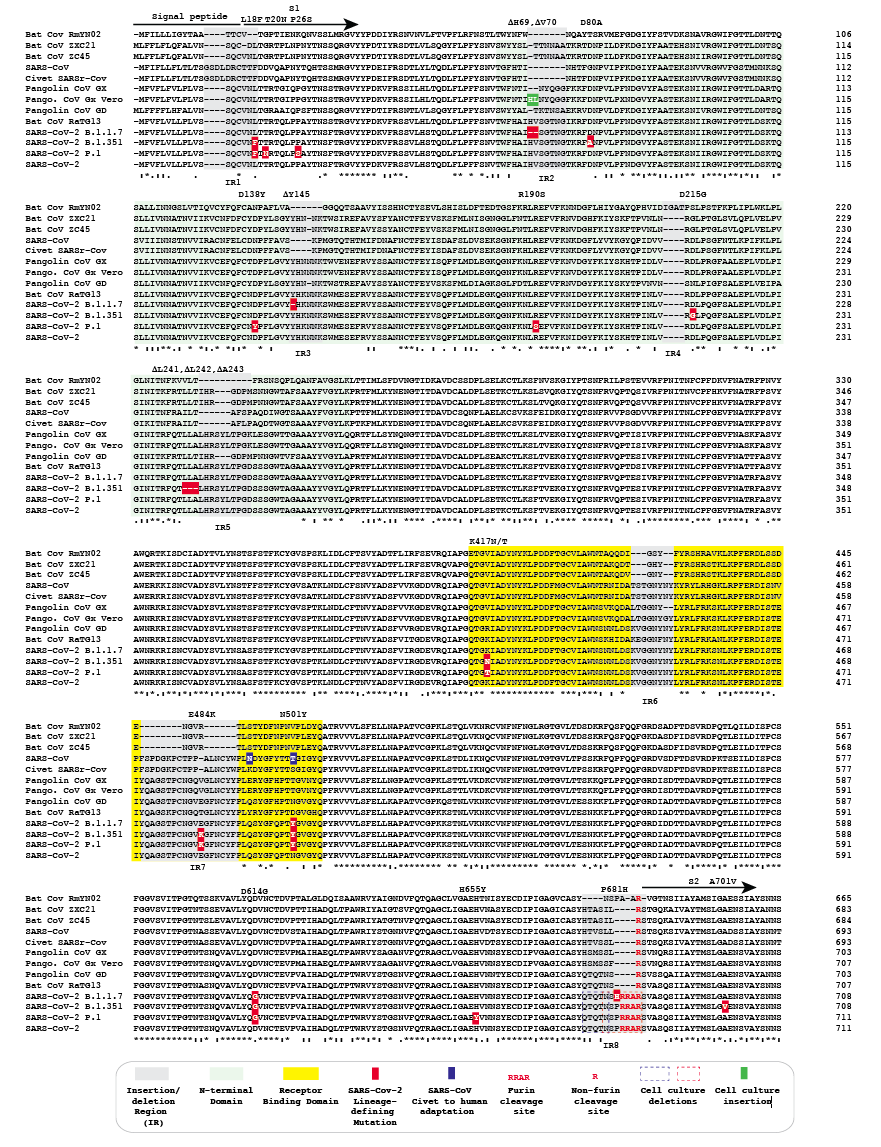
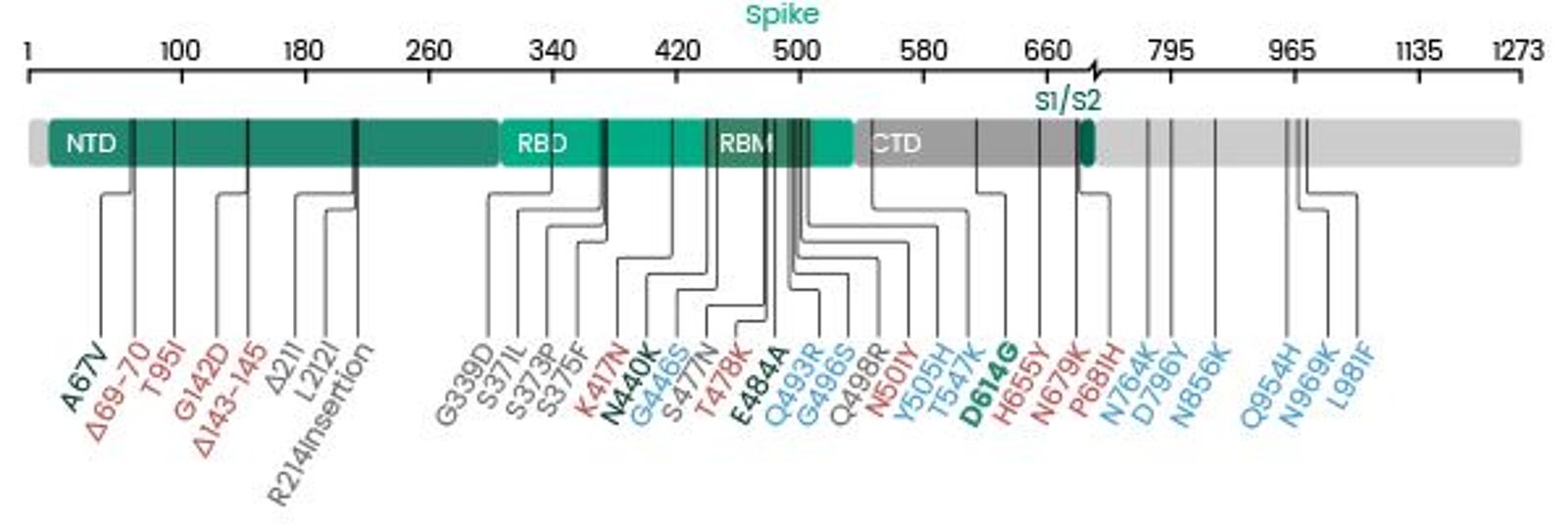
Spike protein mutations in novel SARS-CoV-2 'variants of concern' commonly occur in or near indels - Virological
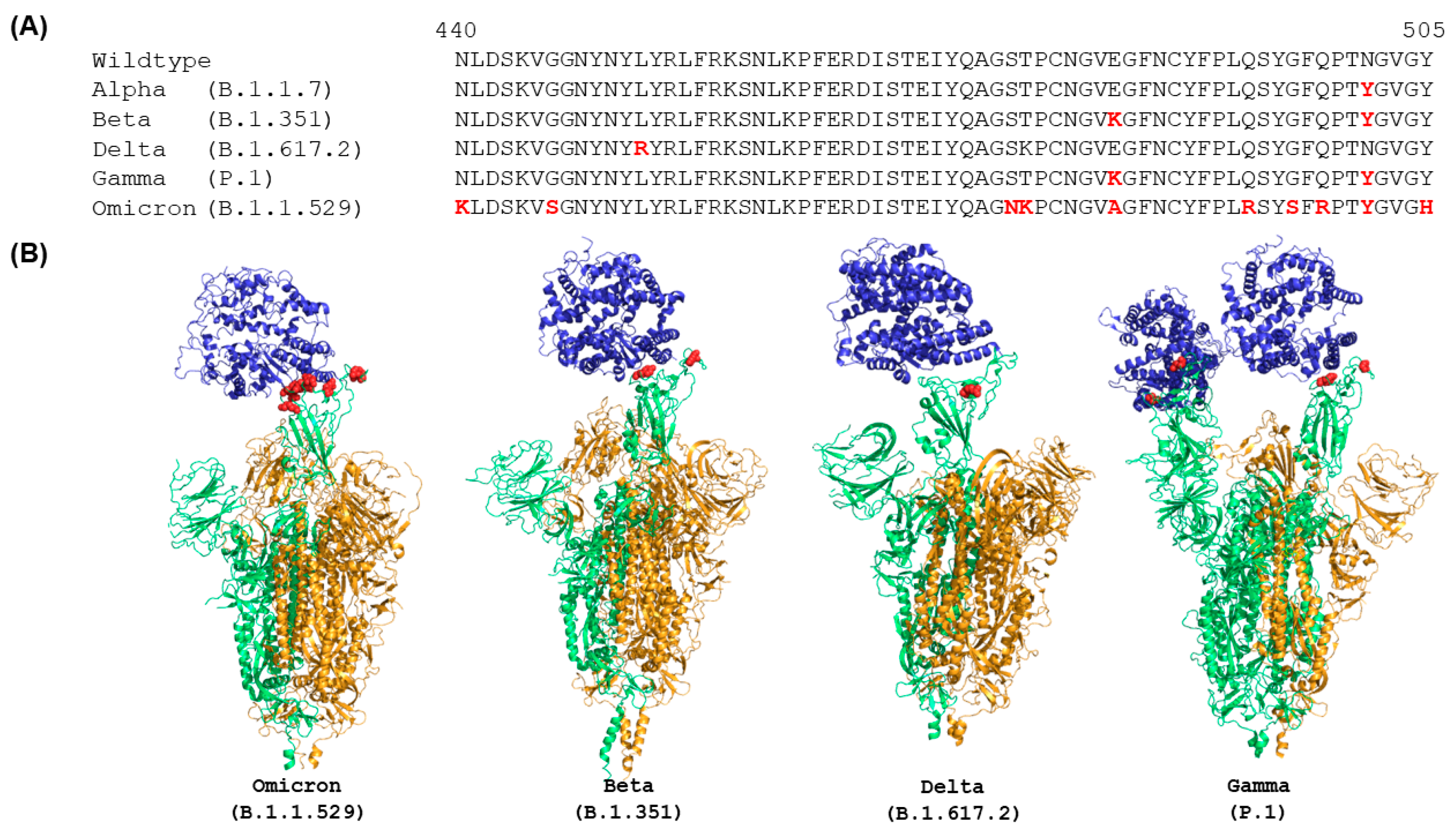
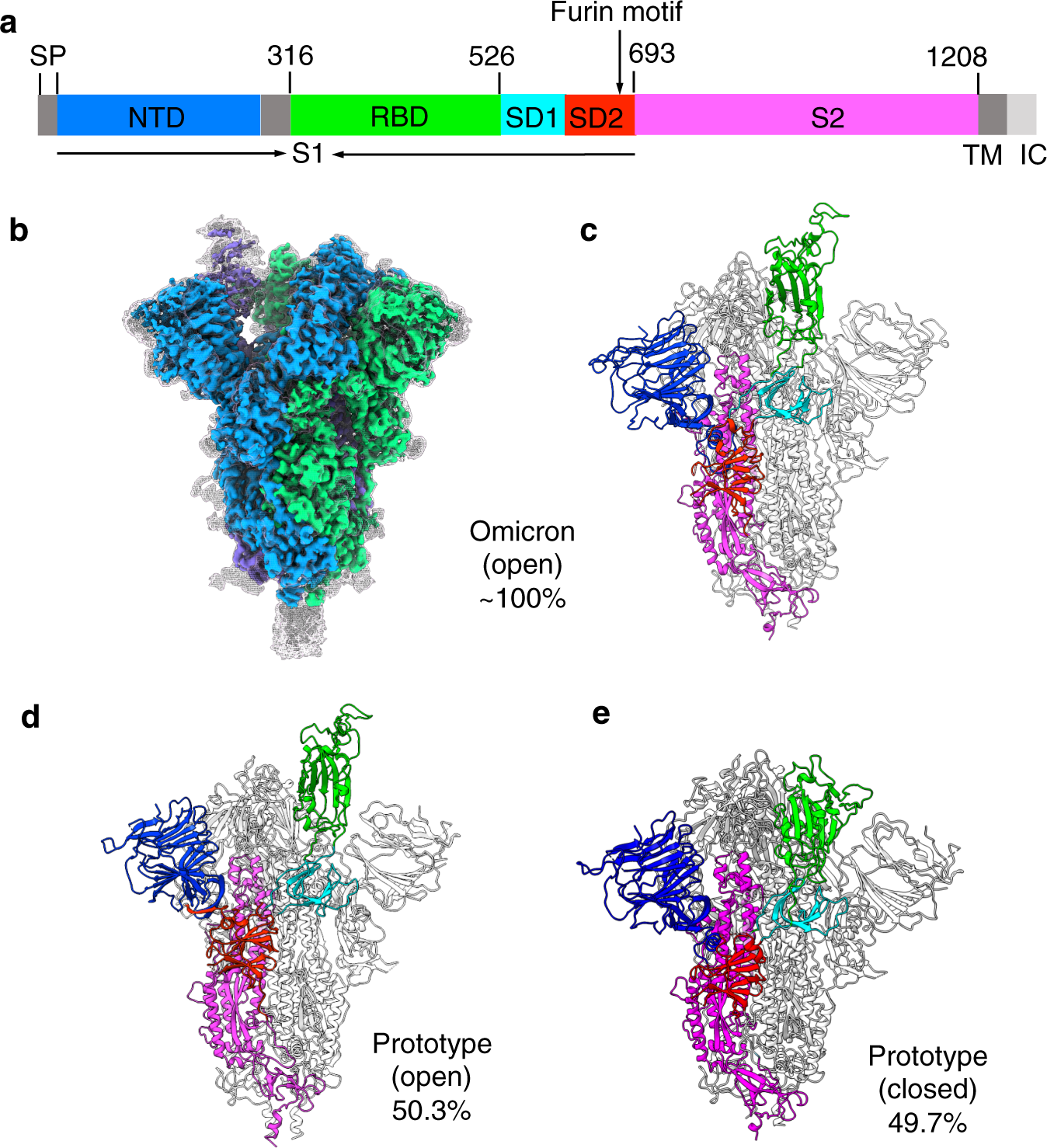
IJMS | Free Full-Text | Distinct Conformations of SARS-CoV-2 Omicron Spike Protein and Its Interaction with ACE2 and Antibody

SARS-CoV-2 (COVID-19) Omicron Variant Spike S1 (His-Avi Tag) Recombinant Protein - Cat. No. 95-129 | ProSci