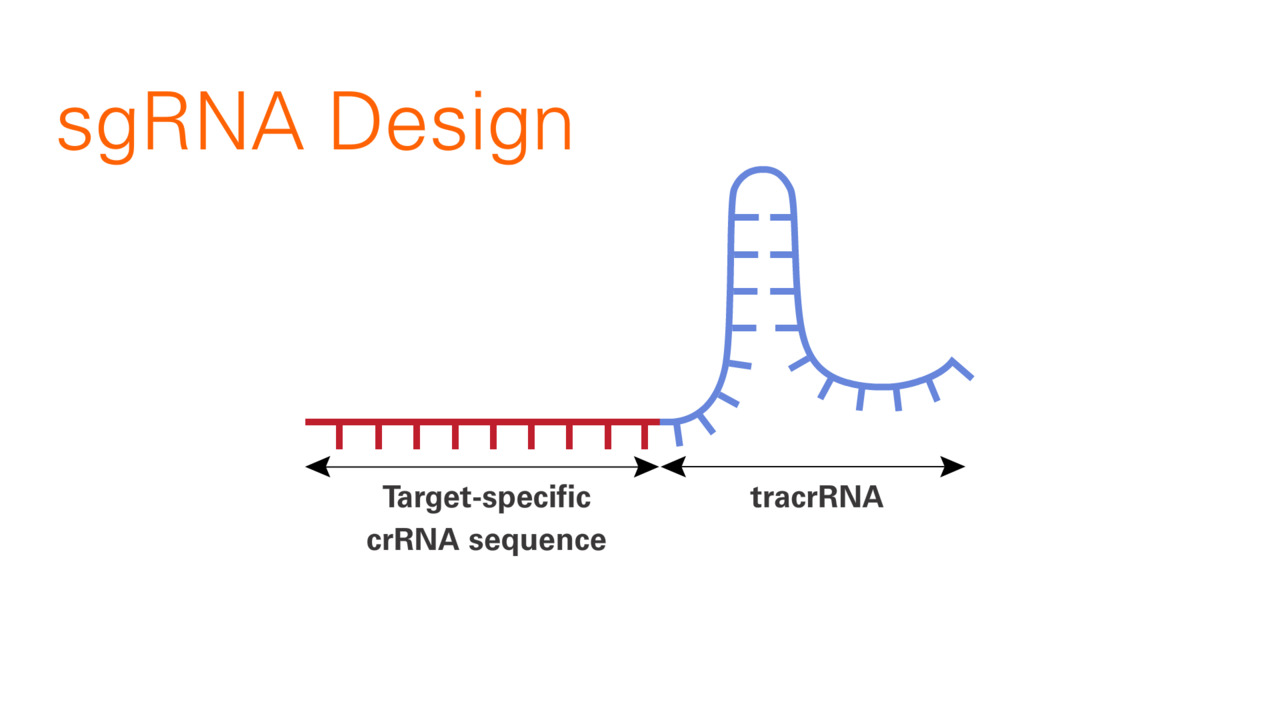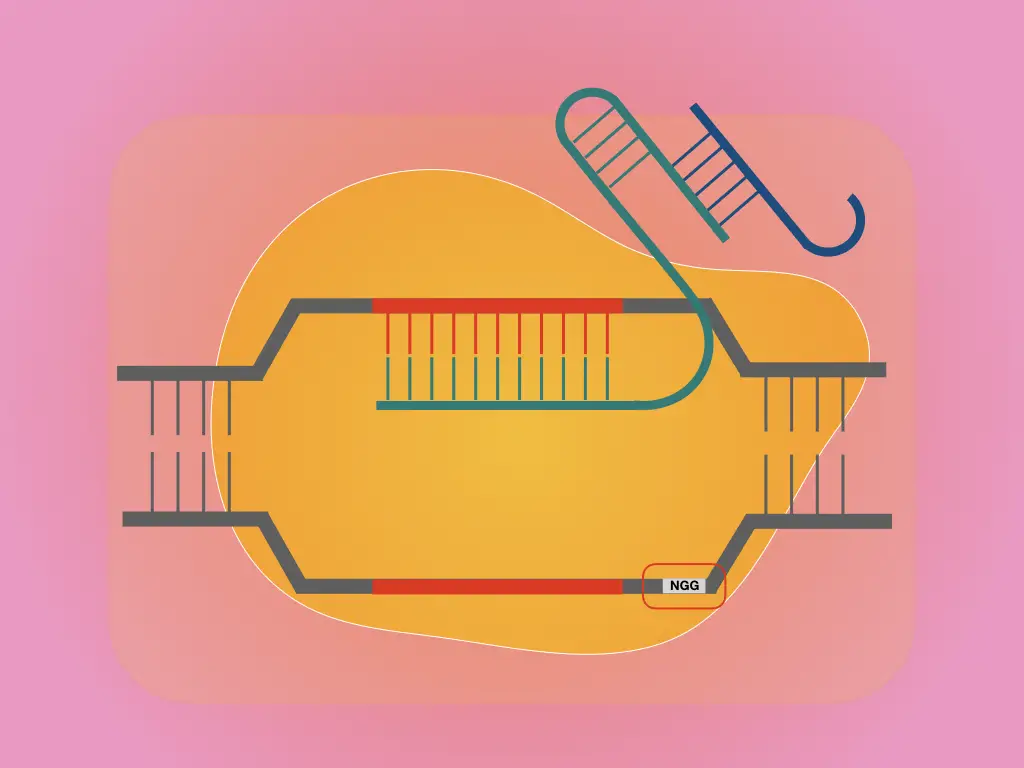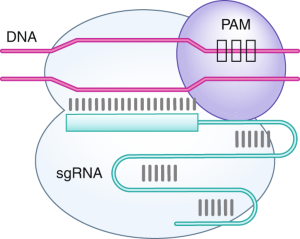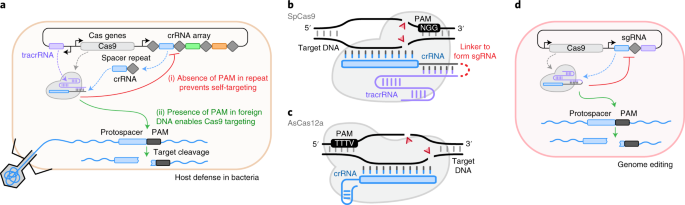
Scalable characterization of the PAM requirements of CRISPR–Cas enzymes using HT-PAMDA | Nature Protocols

Comprehensive PAM prediction for CRISPR-Cas systems reveals evidence for spacer sharing, preferred strand targeting and conserved links with CRISPR repeats | bioRxiv

In Silico Processing of the Complete CRISPR‐Cas Spacer Space for Identification of PAM Sequences - Mendoza - 2018 - Biotechnology Journal - Wiley Online Library
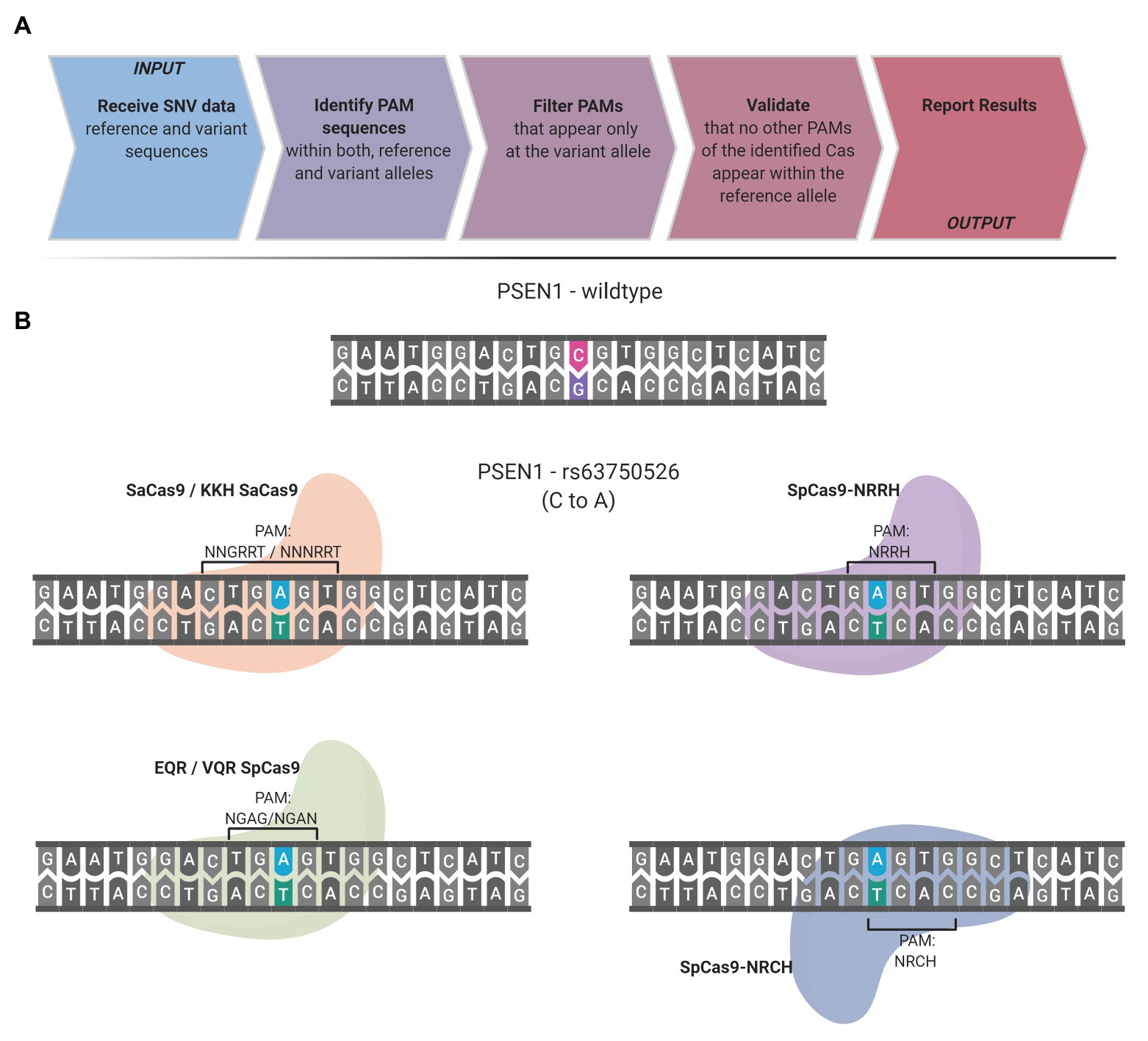
Frontiers | CrisPam: SNP-Derived PAM Analysis Tool for Allele-Specific Targeting of Genetic Variants Using CRISPR-Cas Systems


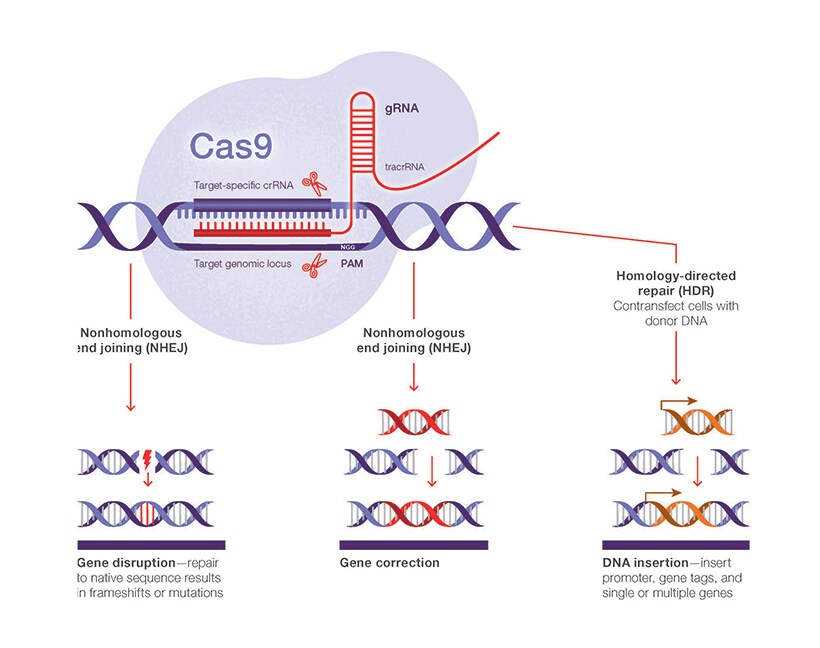




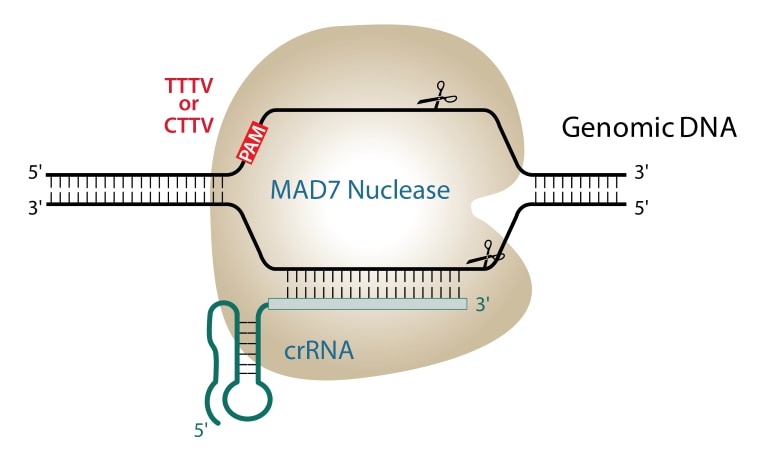
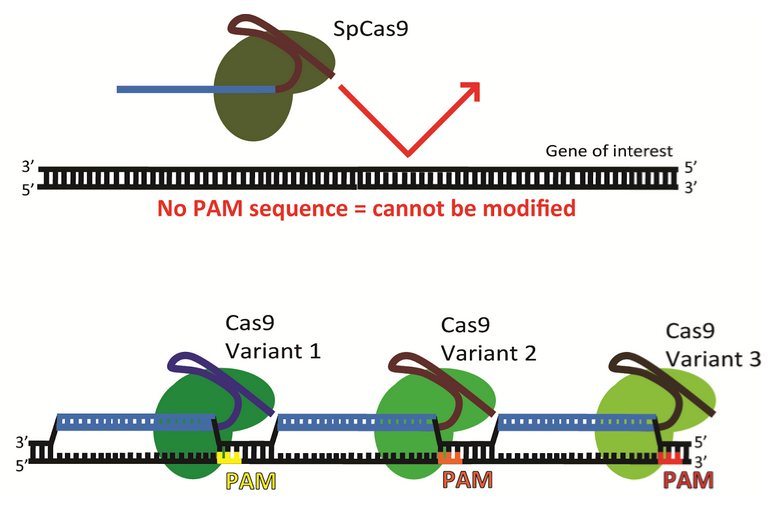


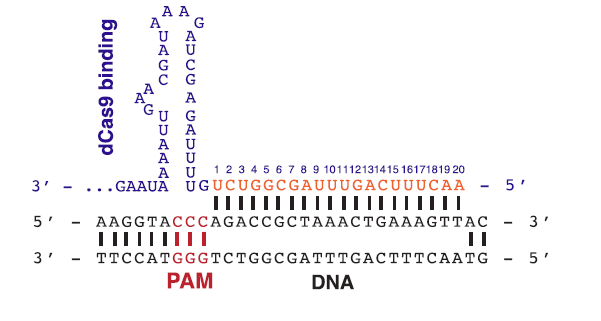

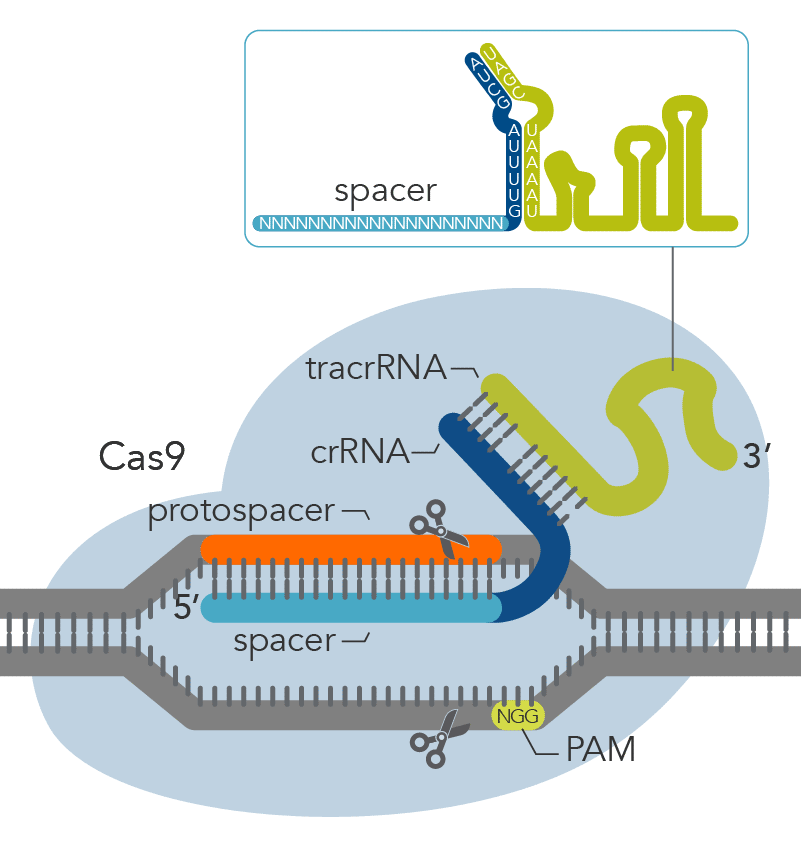


.png?sfvrsn=58c63b07_0)