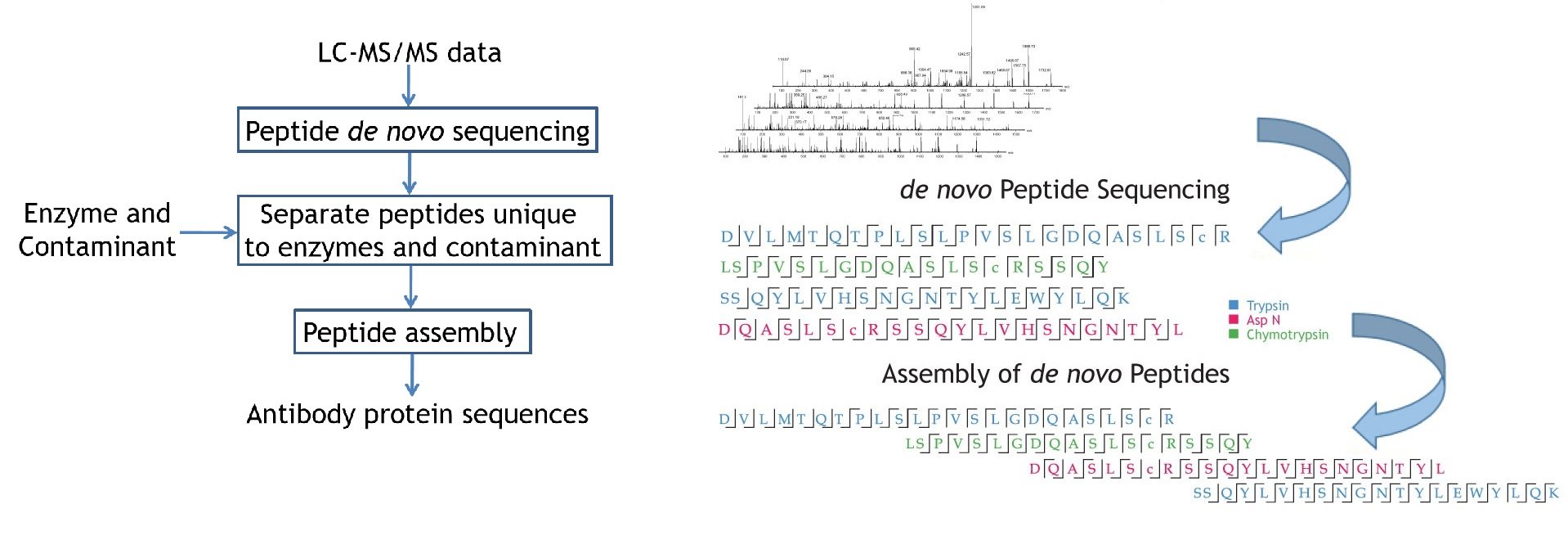
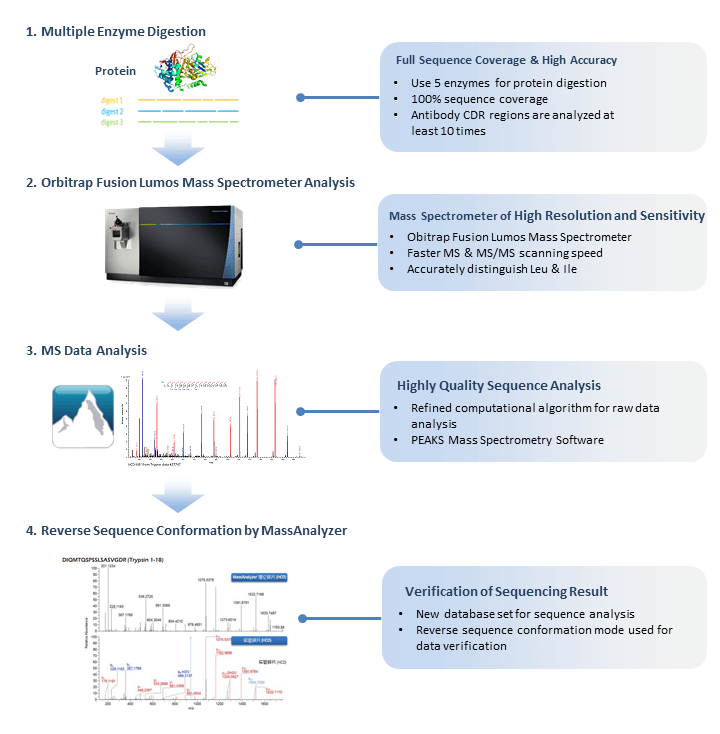
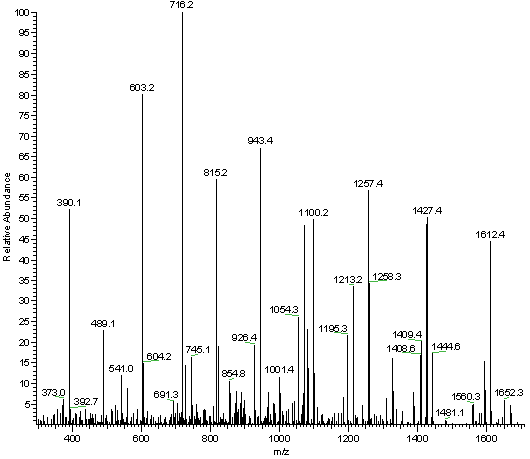
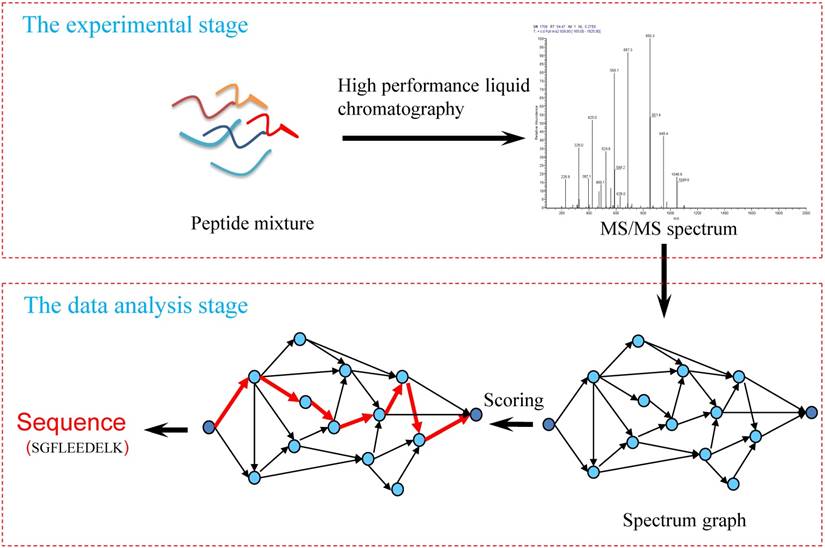
Deep learning enables de novo peptide sequencing from data-independent-acquisition mass spectrometry | Nature Methods

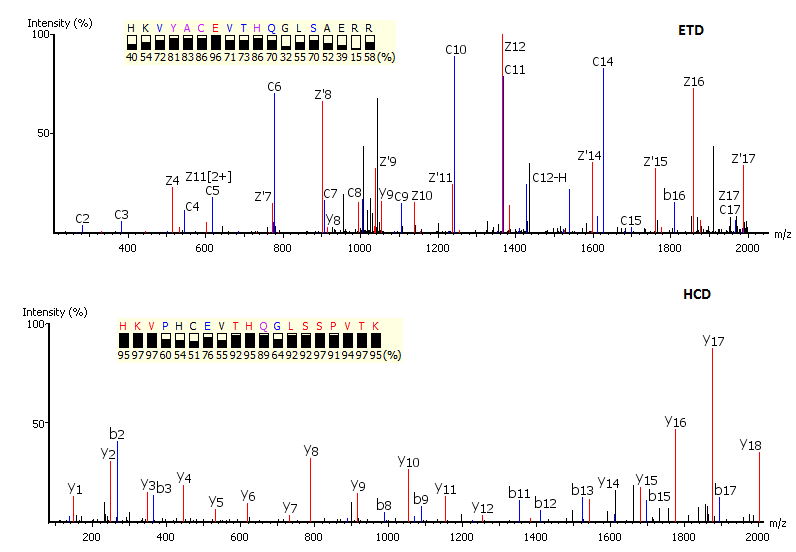
Precision De Novo Peptide Sequencing Using Mirror Proteases of Ac-LysargiNase and Trypsin for Large-scale Proteomics. - Abstract - Europe PMC

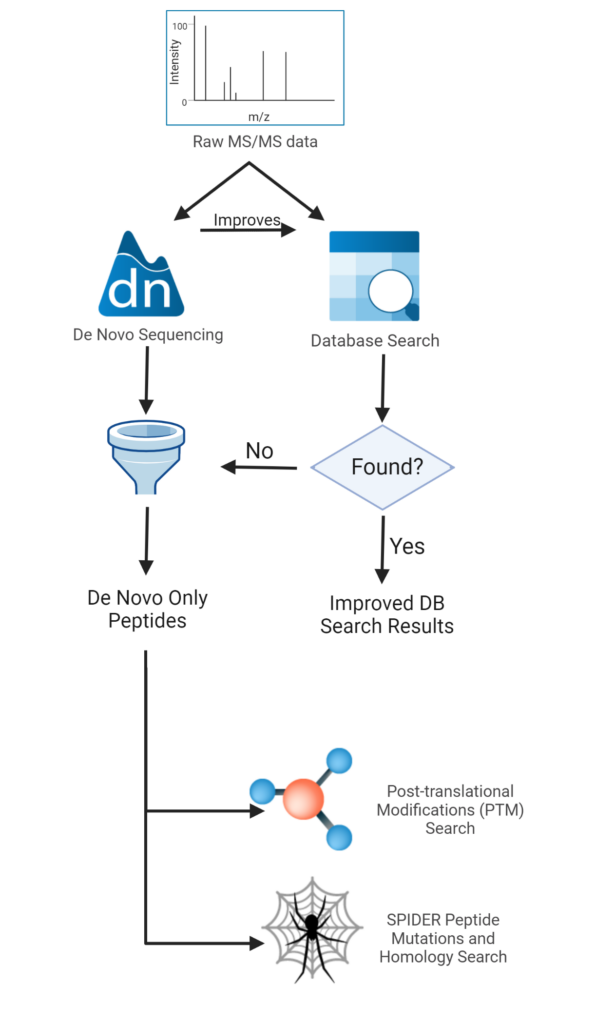
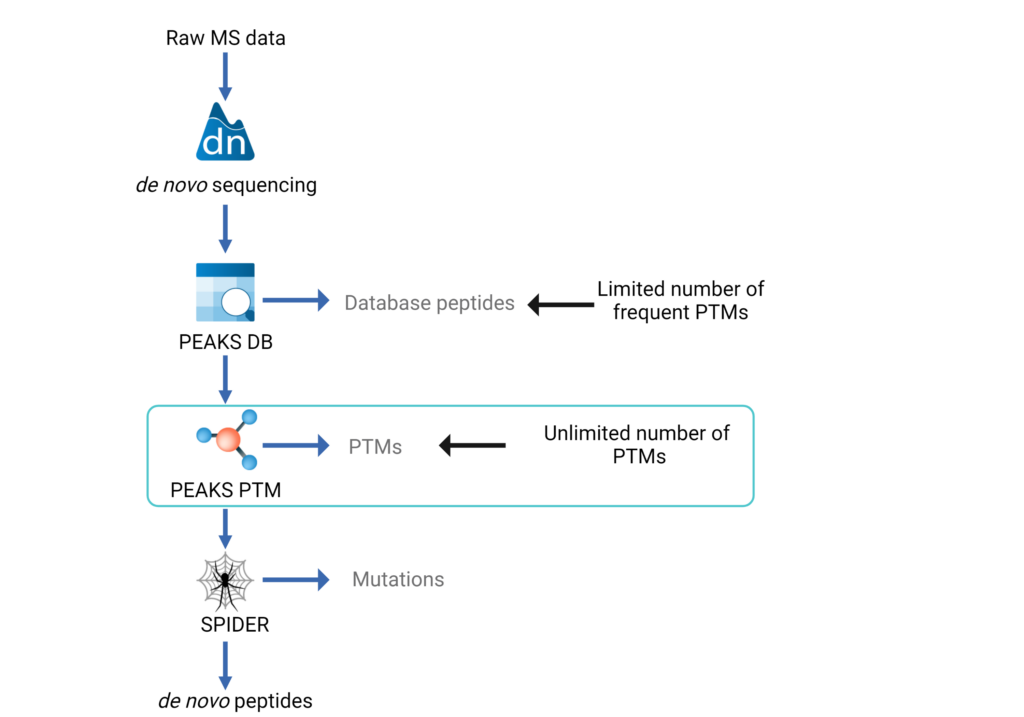
Lookup Peaks: A Hybrid of de Novo Sequencing and Database Search for Protein Identification by Tandem Mass Spectrometry | Analytical Chemistry

Precision De Novo Peptide Sequencing Using Mirror Proteases of Ac-LysargiNase and Trypsin for Large-scale Proteomics - ScienceDirect

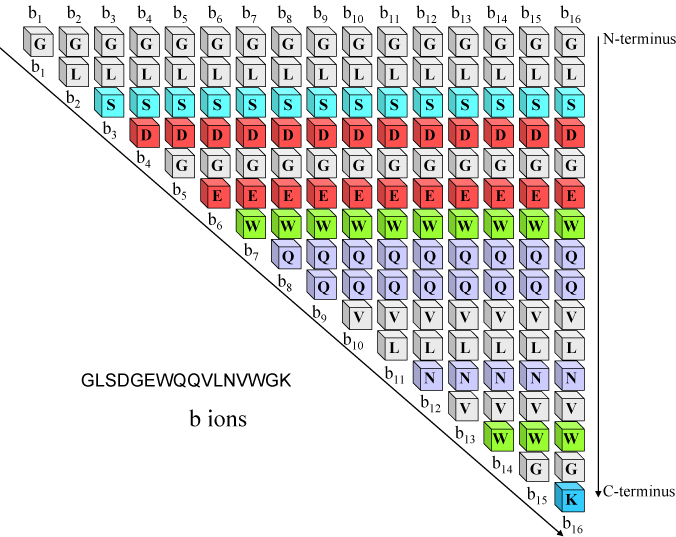
The comparison of de novo sequencing results (PEAKS 5.3) with database... | Download Scientific Diagram

Application of de Novo Sequencing to Large-Scale Complex Proteomics Data Sets | Journal of Proteome Research