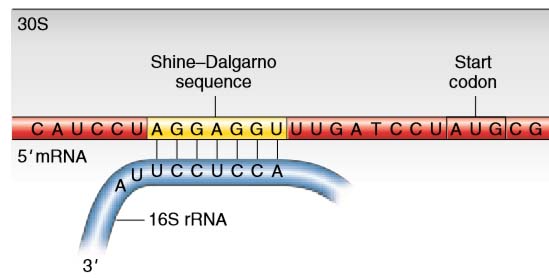
SOLVED: Question 37 2 pts What is the function of the Shine Dalgarno sequence in bacteria? it serves as a spliceosome binding site during post- transcriptional modification it serves as an RNA

Leveraging genome-wide datasets to quantify the functional role of the anti- Shine–Dalgarno sequence in regulating translation efficiency | Open Biology

What is the Difference Between Shine Dalgarno and Kozak Sequence | Compare the Difference Between Similar Terms
Prokaryotic coding regions have little if any specific depletion of Shine- Dalgarno motifs | PLOS ONE

The possible dual impacts of Shine-Dalgarno (SD) sequences on protein... | Download Scientific Diagram

The Shine-Dalgarno sequence of riboswitch-regulated single mRNAs shows ligand-dependent accessibility bursts | Nature Communications

Extending the Spacing between the Shine–Dalgarno Sequence and P-Site Codon Reduces the Rate of mRNA Translocation - ScienceDirect

What is the Difference Between Shine Dalgarno and Kozak Sequence | Compare the Difference Between Similar Terms

Defining the anti-Shine-Dalgarno sequence interaction and quantifying its functional role in regulating translation efficiency | bioRxiv

Accessibility of the Shine-Dalgarno Sequence Dictates N-Terminal Codon Bias in E. coli - ScienceDirect
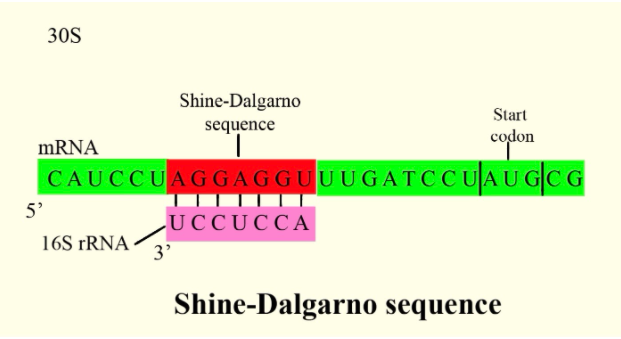
In prokaryotes, the ribosomes binding site on mRNA is called(a)Hogness sequence(b)Shine- Dalgarno sequence(c)Prinbow sequence(d)TATA- box
![PDF] Shine-Dalgarno Anti-Shine-Dalgarno Sequence Interactions and Their Functional Role in Translational Efficiency of Bacteria and Archaea | Semantic Scholar PDF] Shine-Dalgarno Anti-Shine-Dalgarno Sequence Interactions and Their Functional Role in Translational Efficiency of Bacteria and Archaea | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/ca6e3ce301334ffc5d3a7df6b8a8e5d2bc19ed16/56-Table2.1-1.png)