Tracking the amino acid changes of spike proteins across diverse host species of severe acute respiratory syndrome coronavirus 2 - ScienceDirect

Exploring the genomic and proteomic variations of SARS-CoV-2 spike glycoprotein: a computational biology approach | bioRxiv
nCoV-2019 Spike Protein Receptor Binding Domain Shares High Amino Acid Identity With a Coronavirus Recovered from a Pangolin Viral Metagenomic Dataset - nCoV-2019 Evolutionary History - Virological

Wide variabilities identified among spike proteins of SARS Cov2 globally-dominant variant identified | bioRxiv

SARS-CoV-2 Omicron variant: Antibody evasion and cryo-EM structure of spike protein–ACE2 complex | Science
Molecular epidemiology of SARS-CoV-2 clusters caused by asymptomatic cases in Anhui Province, China | BMC Infectious Diseases | Full Text
Spike protein mutations in novel SARS-CoV-2 'variants of concern' commonly occur in or near indels - Virological
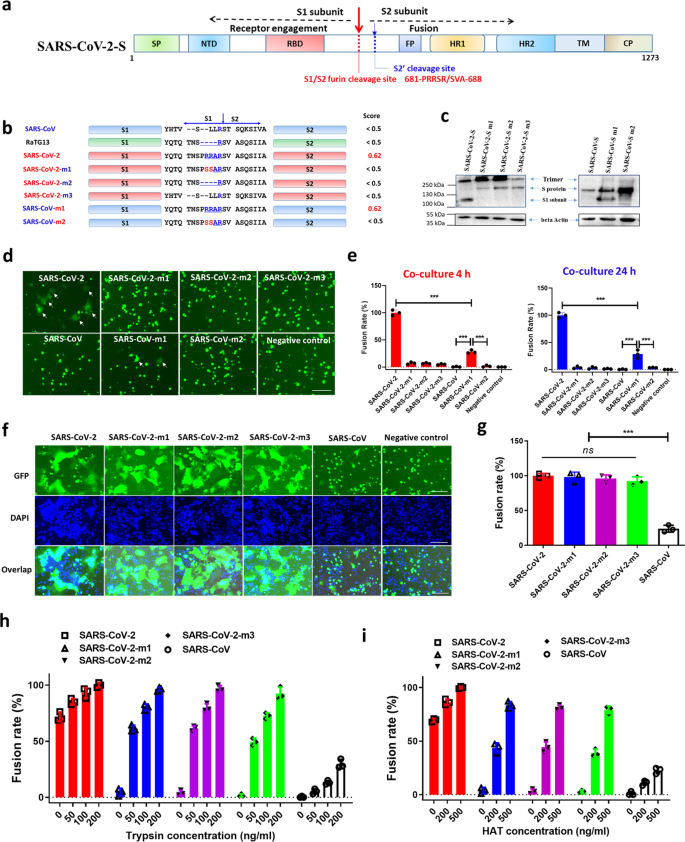
The role of furin cleavage site in SARS-CoV-2 spike protein-mediated membrane fusion in the presence or absence of trypsin | Signal Transduction and Targeted Therapy

Linear B-cell epitopes in the spike and nucleocapsid proteins as markers of SARS-CoV-2 exposure and disease severity - eBioMedicine
Implications of the Mutations in the Spike Protein of the Omicron Variant of Concern (VoC) of SARS-CoV-2 – Signature Science
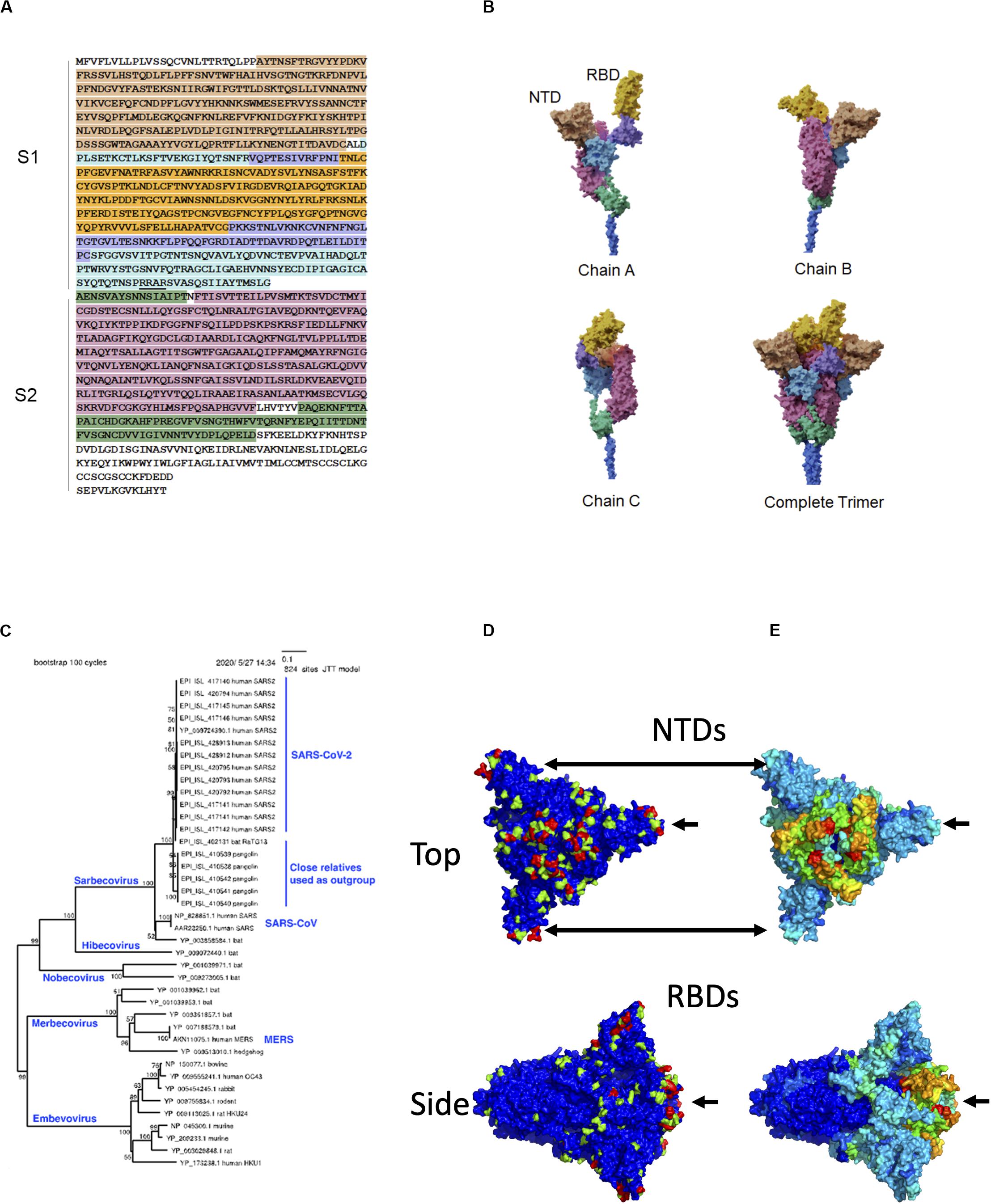
Frontiers | Flexible, Functional, and Familiar: Characteristics of SARS-CoV-2 Spike Protein Evolution

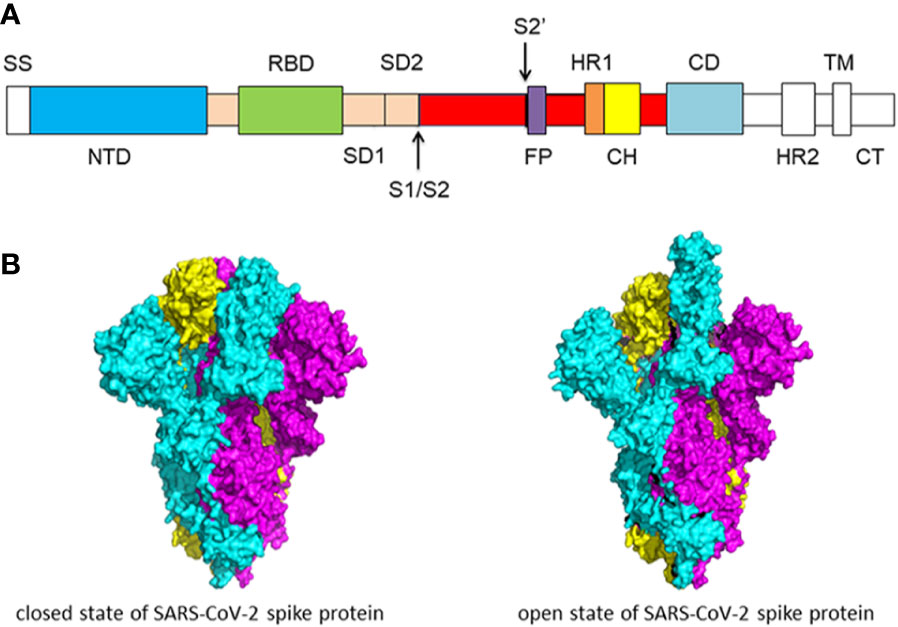
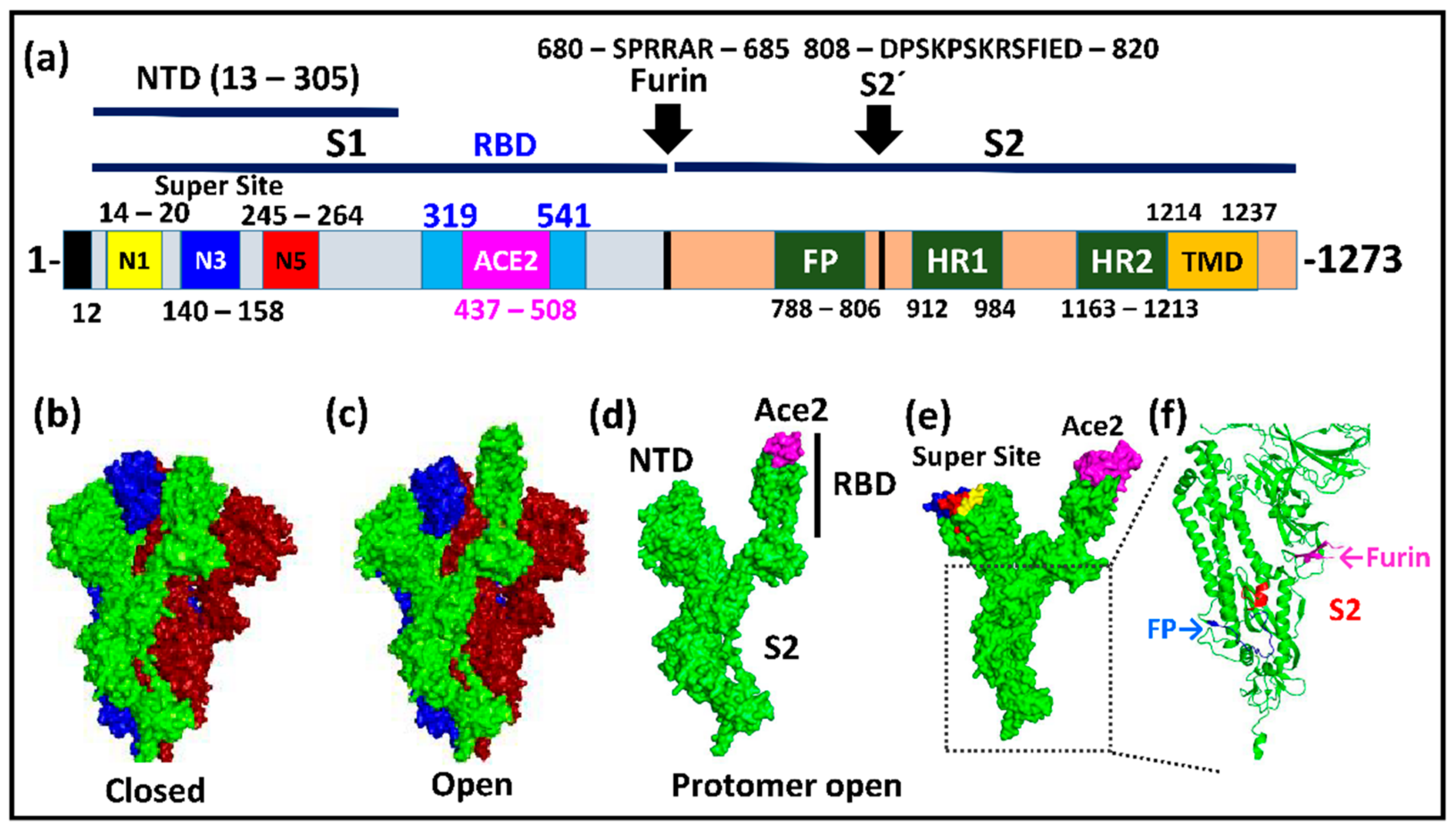
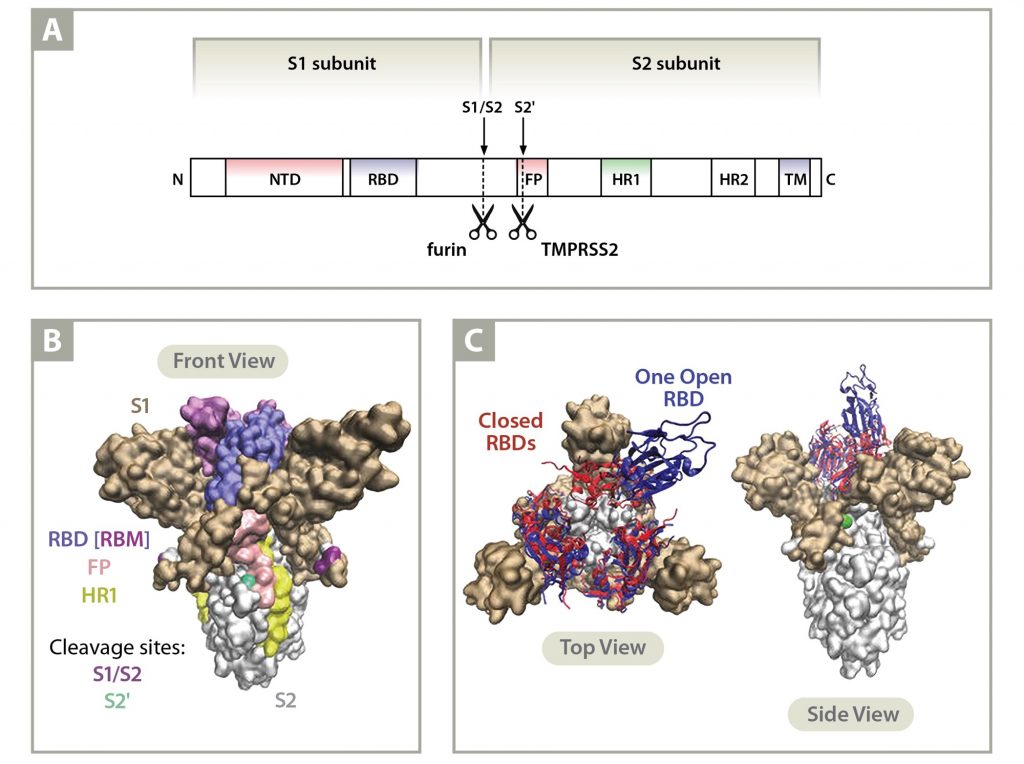
Molecular biology of the SARs‐CoV‐2 spike protein: A review of current knowledge - Zhu - 2021 - Journal of Medical Virology - Wiley Online Library

Identification of Two Critical Amino Acid Residues of the Severe Acute Respiratory Syndrome Coronavirus Spike Protein for Its Variation in Zoonotic Tropism Transition via a Double Substitution Strategy - ScienceDirect