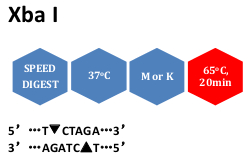
In Vitro Seamless Stack Enzymatic Assembly of DNA Molecules Based on a Strategy Involving Splicing of Restriction Sites | Scientific Reports
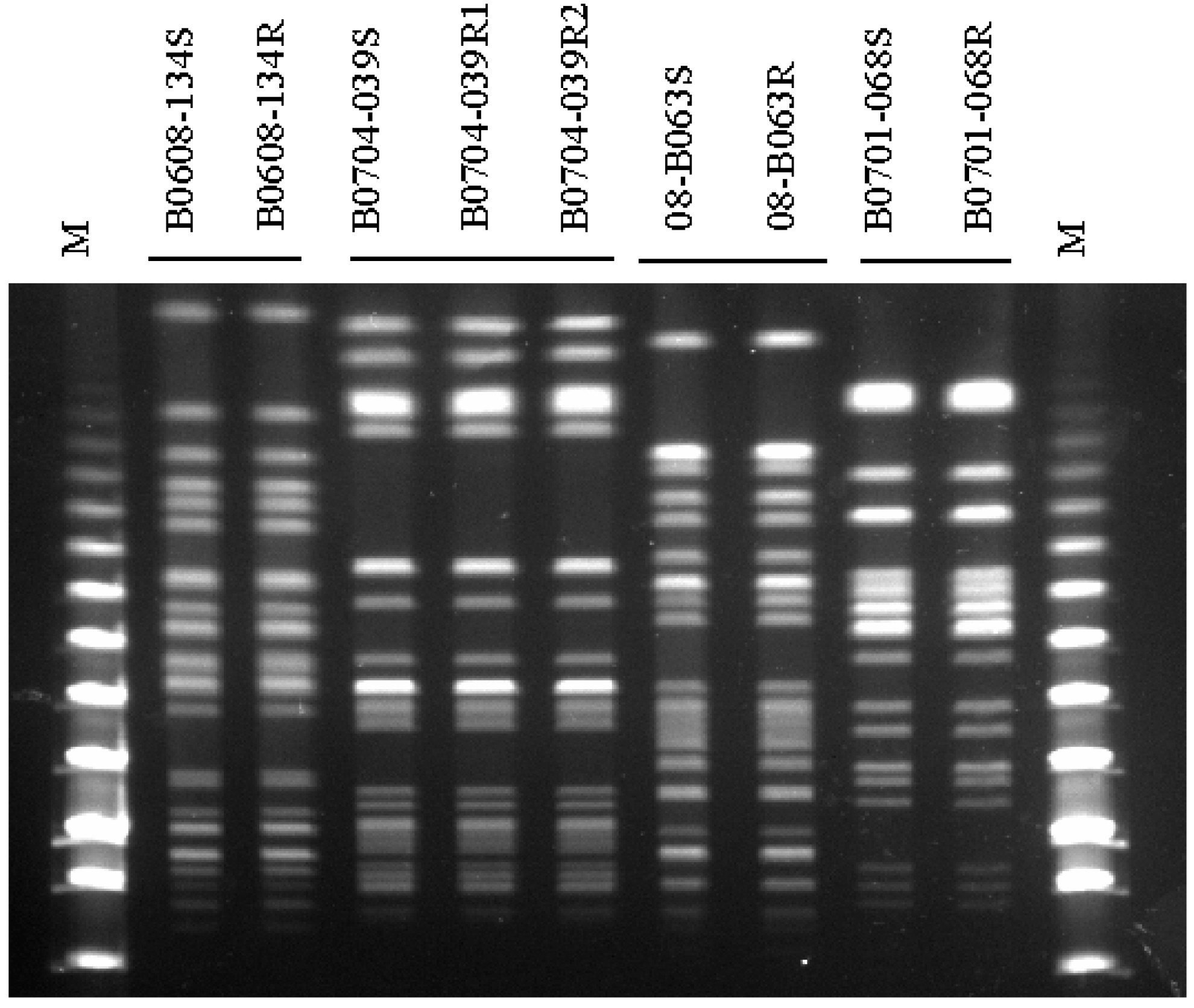
XbaI, PstI, and BglII restriction enzyme maps of the two orientations of the varicella-zoster virus genome. - Abstract - Europe PMC
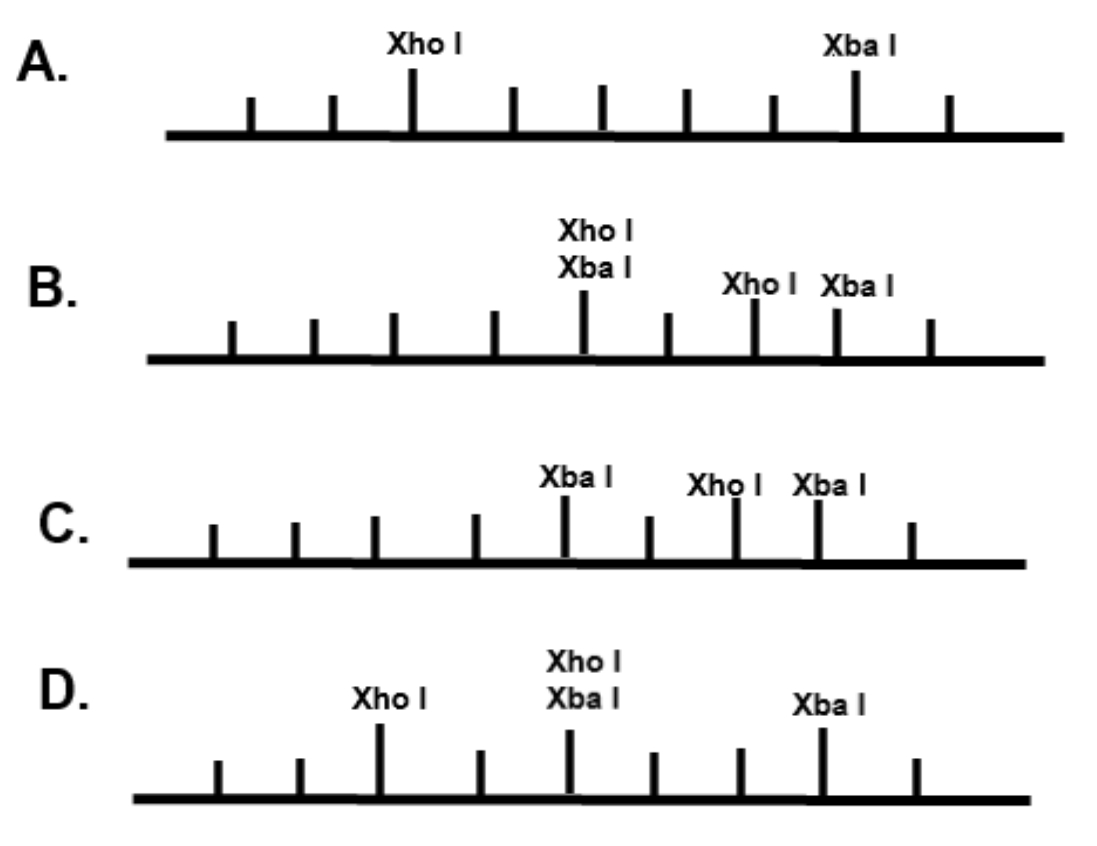
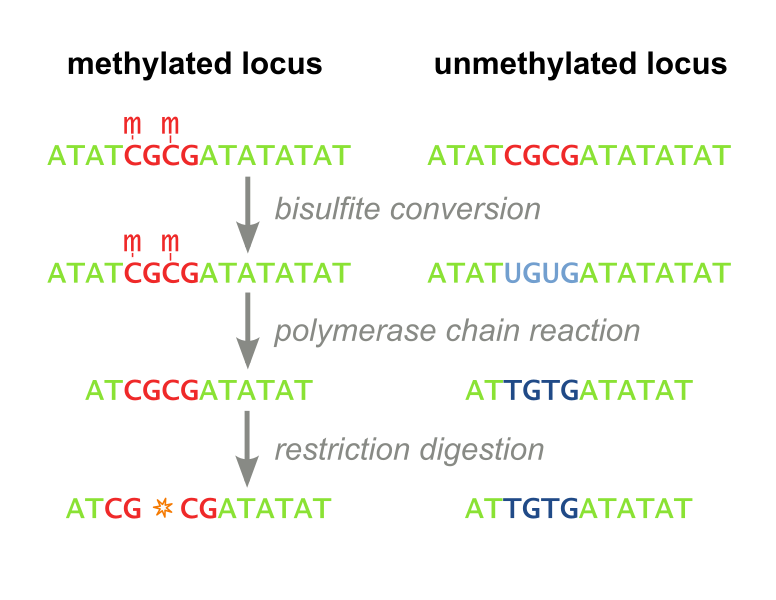
The restriction enzyme XhoI and XbaI were used to digest a linear fragment of DNA at a specific sequence such as that shown for EcoR1 (figure above). Use the information gained from